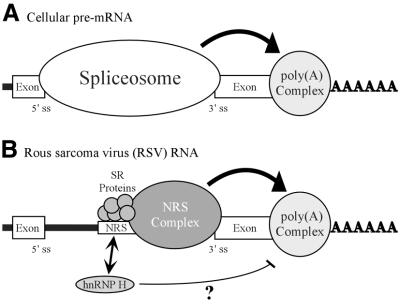
Figure 8.
Model for NRS-mediated stimulation of RSV polyadenylation. (A) Spliceosome contribution to polyadenylation. A generic cellular pre-mRNA is shown with the 5′ and 3′ splice sites of a terminal intron indicated. In cellular RNA processing the terminal 3′ splice site and the polyadenylation site define the last exon and factors associated with the spliceosome act to stimulate polyadenylation (large arrow; see Discussion). (B) NRS contribution to RSV polyadenylation. The NRS is shown between authentic viral splice sites. Depicted are SR proteins that bind to nt 701–770 and initiate NRS complex formation. The spliceosome-like inhibitory complex (NRS complex) that forms between the NRS and a viral 3′ splice site is indicated. The NRS may serve to stabilize the binding of splicing factors to the weak viral 3′ splice site, which can then either recruit or stabilize the polyadenylation complex (large arrow) and thereby enhance polyadenylation of viral unspliced RNA. A potential competition between SR proteins and hnRNP H for binding to this region is reflected by an arrow. hnRNP H binding may be restricted when SR proteins are present. In contrast, if SR proteins are absent or limiting, hnRNP H binding may increase and in some way disrupt the ability of the NRS to act efficiently on the polyadenylation complex (thin line). In both cases the NRS complex is competent for splicing inhibition.