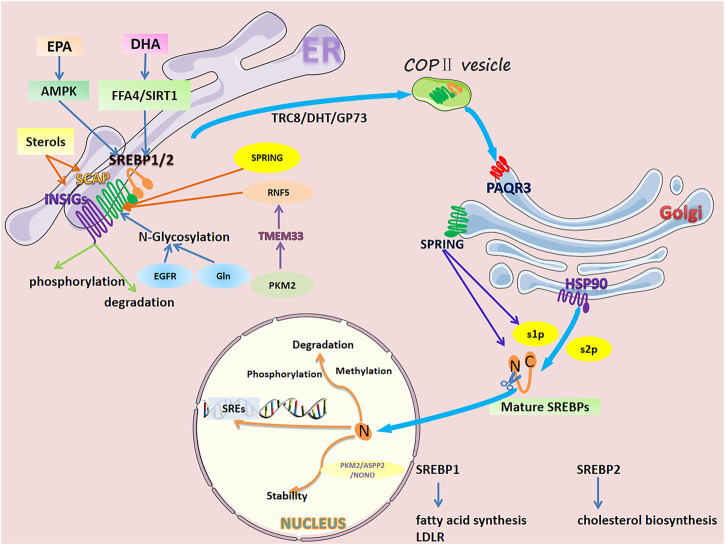
FIGURE 1.
Regulation of SREBP1/2 in cancer cells. The activation process of SREBPs is as follows. Inactive SREBPs reside in the ER membrane and interact with SCAP. The N-terminal domain of SCAP combines with INSIG, forming an INSIG/SCAP/SREBP complex anchored to the ER. When sterol levels decrease, SCAP dissociates from INSIGs and mediates SREBPs into COPII vesicles, transporting the SCAP/SREBP complex from the ER to the golgi. In the golgi, SREBPs are sequentially cleaved by S1P and S2P, releasing their transcriptionally active N-terminal domains. After cleavage, mature SREBPs translocate to the nucleus and bind to SREs and E-boxes within target gene promoters. However, SREBPs are delicately and complexly regulated in individual organelles. In the ER, sterol levels directly affect the dissociation of SCAP from INSIGs. Long-chain polyunsaturated fatty acids (DHA and EPA) inhibit SREBPs at the mRNA and protein levels. N-glycosylation of SCAP, RNF5-induced degradation, SPRING-induced reduction, phosphorylation, and degradation of INSIGs all affect the transport of SREBPs to the golgi. In the golgi, PAQR3 promotes SCAP/SREBP localization and enhances the processing of SREBPs. HSP90 binds the SREBP-SCAP complex, stabilizing it and facilitating its transport from the ER to the golgi. SPRING, a necessary cofactor for the cleavage of SREBPs, directly affects the level of SREBP. In the nucleus, mature SREBPs undergo phosphorylation, methylation, and ubiquitination-related degradation. Additionally, protein-protein interactions affect their stability.