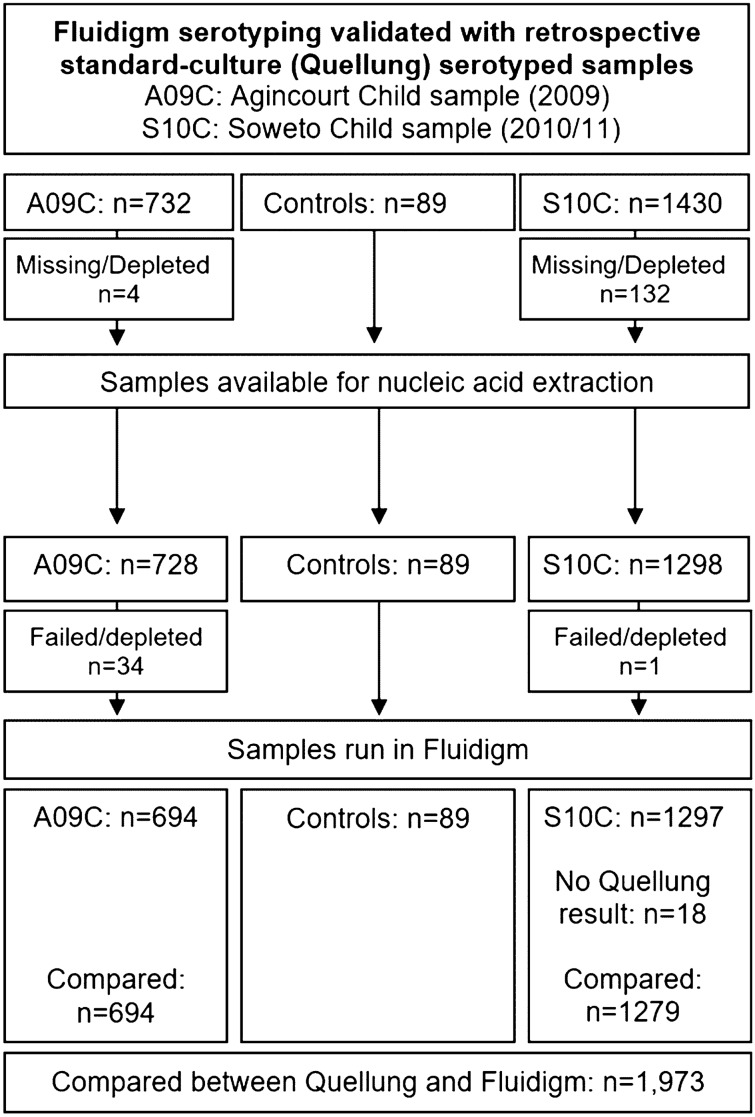
Figure 1.
Validation flowchart describing the included archived clinical samples that were previously serotyped using the culture based Quellung method and that were compared with the Standard BioTools ‘Fluidigm’ real-time quantitative and serotyping PCR. A09C indicates samples collected from Agincourt (2009) children39. S10C indicates samples from Soweto (2010/11) children40. Samples were archived nasopharyngeal swabs in STGG as described in "Clinical samples (nasopharyngeal swabs) for validation" section, that were previously tested using the culture based Quellung method39,40. Archived clinical samples were used to assess the diagnostic performance of the ‘Fluidigm’ qPCR. Where samples were indicated as missing/depleted, these samples were not available for nucleic acid extraction as they were either exhausted through previous testing, or the sample vial was not located. Where samples are indicated as failed/depleted, the nucleic acid extraction failed and there was insufficient remaining sample to repeat this extraction. Control samples were culture strains used to assess the analytical performance of the ‘Fluidigm’ qPCR and are described in methods "Total nucleic acid extraction" section. All extracted samples and controls were run in the ‘Fluidigm’, however only samples with a Quellung result (positive or negative) were included in comparisons of diagnostic performance.