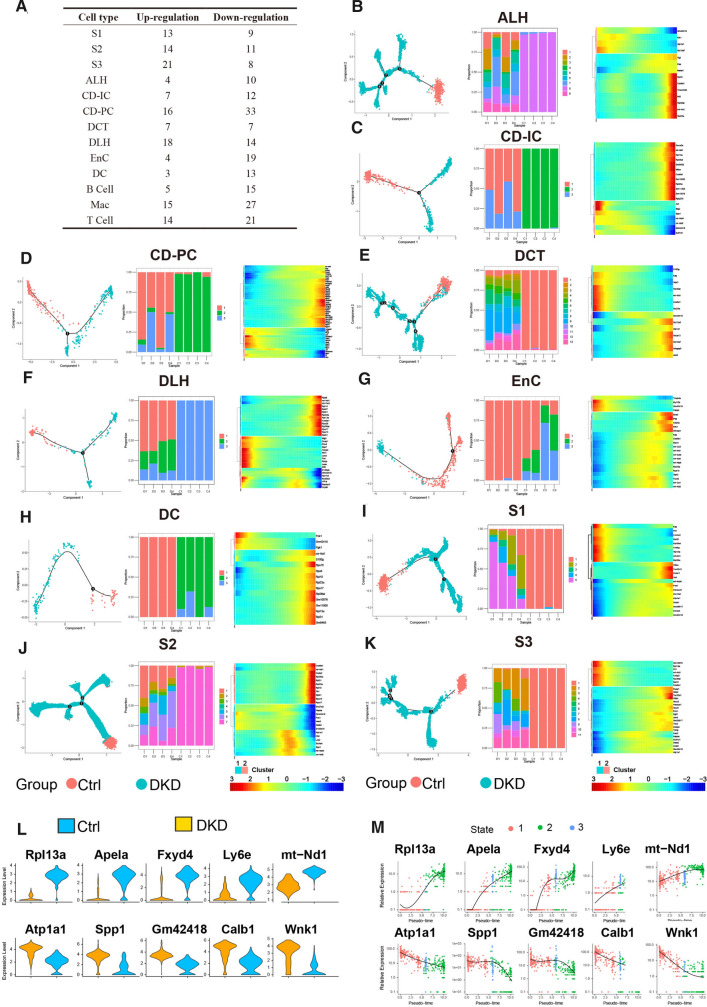
Fig. 4.
The cells of DKD group and Ctrl group showed significant differences in Pseudotime analysis. A. The table recorded the distribution of both up-regulated and down-regulated DEGs in all renal cells. B-K. Trajectory inference obtained by Monocle using RNA velocities method. These graphs contain three forms of results severally. The first being, cells were displayed in trajectory dimensionality reduction colored by group. Second, distributions of renal stromal cells in different states among each sample were shown in a percentage bar chart respectively. Third, the heatmap showed gene expression (evaluated by Z-value) as the transition during time dynamics of pseudotime. L. Top5 up-regulated DEGs and down-regulated DEGs were selected according to the absolute value of fold change generated by the comparison of expression levels between group DKD and group Ctrl in CD-PC. M. Expression of DEGs in Fig. 4E varied during transition with pseudotime states of CD-PC