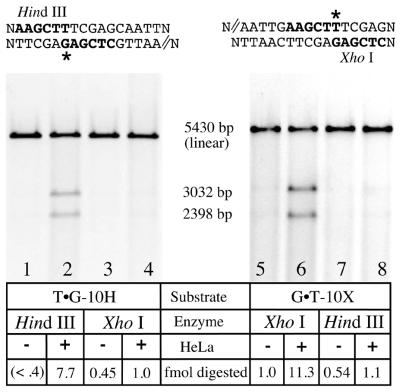
Figure 5.
Strand-specific processing of G·T mismatches. Heteroduplexes (80 ng, 22.5 fmol) containing a G·T mismatch and a 5′-nick placed 10 bp away in either strand were assayed for mismatch correction in each strand. All molecules were linearized with StyI or AvaI, which cuts at a site remote from the mismatch (not shown). In the absence of HeLa treatment (odd numbered lanes) neither scoring enzyme (HindIII or XhoI; refer to the sequence information above each gel) substantially digested at the mismatch site; linear molecules are primarily observed. When the substrates were incubated with HeLa nuclear extract for 20 min under assay conditions (Materials and Methods), the base located in the nicked strand (indicated by double lines in the sequence above the gels) was partially excised and replaced. Resynthesis through this site restores HindIII or XhoI sensitivity, depending on the substrate (lanes 2 and 6) that was used to quantitate strand-specific MR (7,10). For the T·G-10H substrate (left), 35% (7.7 fmol) of the DNA is repaired, whereas for the G·T-10X, 50% of the molecules (11.3 fmol) are repaired. The putatively continuous strand remains largely insensitive to such treatment (lanes 4 and 8).