Figure 4.
STMN2 mis-splicing can be rescued in 2D
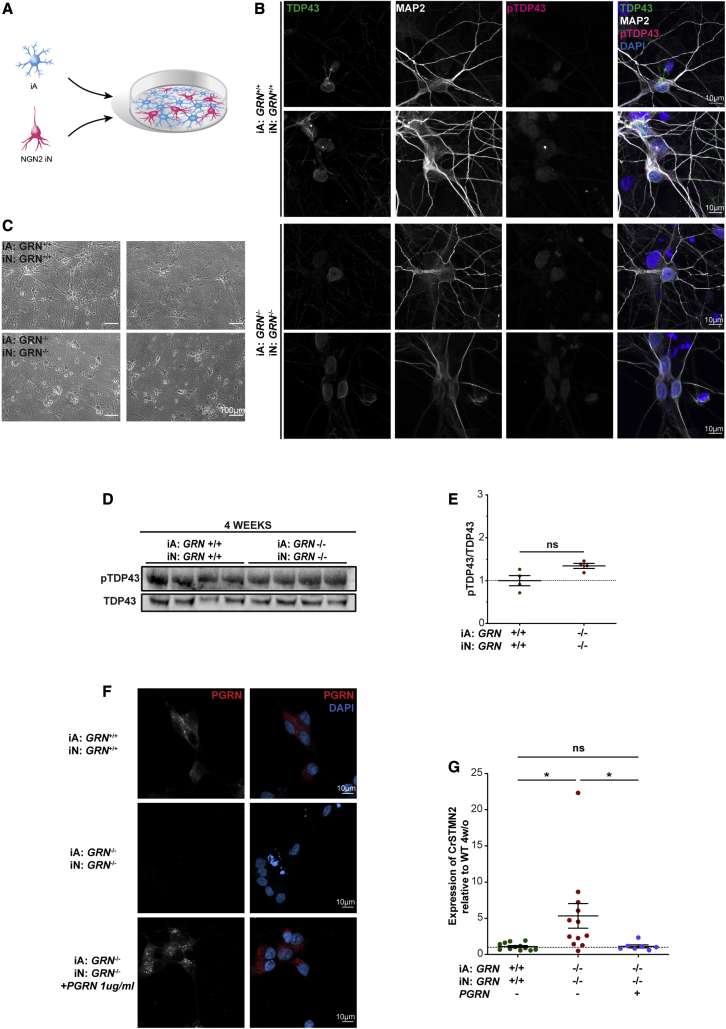
(A) Diagram showing the process by which 2D cultures are made. Briefly, iPSCs are differentiated into mature cortical-like astrocytes and neurons and assembled in a 1:1 ratio in 2D co-cultures. The co-cultures are grown for 4 weeks and then analyzed.
(B) Representative ICC images of 2D cultures stained for TDP-43, pTDP-43, MAP2, and DAPI (scale bar, 10 μm).
(C) Brightfield image of 2D cultures (GRN+/+, GRN−/−) at the 4-week timepoint (scale bar, 100 μm).
(D) Western blot of 2D co-cultures whole lysate of pTDP-43 and total TDP-43 in GRN+/+ and GRN−/− 2D co-cultures.
(E) Western blot quantification showing similar expression on pTDP-43 in GRN−/− compared with GRN+/+ 2D co-cultures when normalized to total TDP-43 (n = 4, unpaired t test, two tailed, p > 0.05, each n represents approximately 1 × 106 cells and was repeated independently four times).
(F) Representative ICC images of 2D cultures stained for PGRN and DAPI showing positive staining in both GRN+/+ and PGRN treated GRN−/−but no PGRN staining in GRN−/− co-cultures (scale bar, 10 μm).
(G) Quantification of CrSTMN2 expression using qPCR showing significantly higher expression in GRN−/− compared with GRN+/+ 2D co-cultures. The difference is rescued when GRN−/− are treated with PGRN for 4 weeks (n = 10, one-way ANOVA followed by multiple comparison, ∗p < 0.05, ns p > 0.05, each n represents approximately 1 × 106 cells and was repeated independently three times). For all graphs, data are presented as mean ± standard error of the mean.