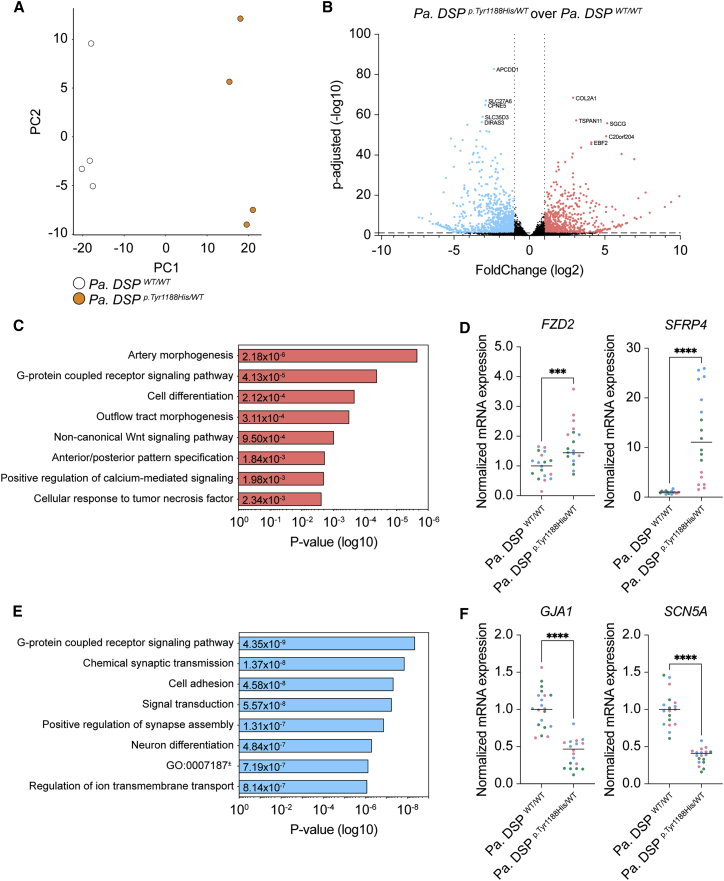
Figure 3.
Gene networks related to Wnt signaling and ion handling are dysregulated in heterozygous Pa. DSPp.Tyr1188His/WT hiPSC-derived cardiomyocytes
(A–F) mRNA sequencing analyses on 1-month-old hiPSC-derived cardiomyocytes.
(A) Principal-component analysis (PCA) for Pa. DSPWT/WT and Pa. DSPp.Tyr1188His/WT cardiomyocytes.
(B) Volcano plot showing the up- and downregulated genes (fold change (log2) >1 and <−1; p-adj < 0.05) in Pa. DSPp.Tyr1188His/WT and control cardiomyocytes. The top five up- and downregulated genes are indicated.
(C) Gene Ontology analysis on the upregulated (fold change (log2) >1) genes.
(D) Validation of FZD2 and SFRP4, both belonging to the “non-canonical Wnt-signaling pathway” term.
(E) Gene Ontology analysis on the downregulated (fold change (log2) <−1) genes.
(F) Validation of GJA1 and SCN5A, both belonging to the “regulation of ion transmembrane transport” term. mRNA sequencing was performed on four replicates obtained from one hiPSC differentiation.
The dots in (D) and (F) represent technical replicates, whereas the color of each dot indicates the experimental origin (3 independent experiments; 4–8 technical replicates). Gene expression data are normalized to GUS and plotted as mean. Significance has been assessed by a two-tailed unpaired Student’s t test (∗∗∗p < 0.001, ∗∗∗∗p < 0.0001). GO: 0007187, G protein-coupled receptor signaling pathway coupled to cyclic nucleotide second messenger.