Figure 4.
hiPSC-derived KI. DSPp.Tyr1188His/WT cardiomyocytes corroborate findings observed in patient-derived cardiomyocytes
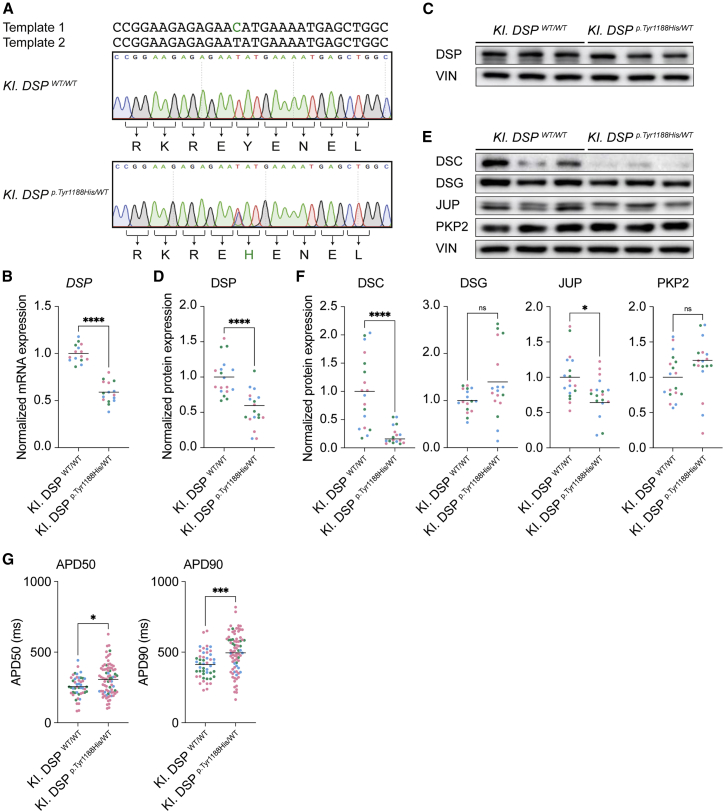
(A) Sanger sequencing traces of the control and knockin hiPSC lines. The two different DNA templates used to introduce the intended mutation are depicted. Intended mutation is indicated in green.
(B–G) Molecular and functional analyses on 1-month-old hiPSC-derived cardiomyocytes obtained from three independent experiments.
(B) Gene expression levels of DSP normalized to GUS.
(C) Representative immunoblots for DSP.
(D) Quantification of DSP protein levels normalized to VIN.
(E) Representative immunoblots for DSC, DSG, JUP, and PKP2.
(F) Quantification of the desmosomal protein levels. Values normalized to VIN.
(G) APD measured at 50% and 90% (KI. DSPWT/WT, n = 48 cell clusters; KI. DSPp.Tyr1188His/WT, n = 81 cell clusters) of cardiomyocyte repolarization.
Data are plotted as mean. The dots in (B), (D), (F), and (G) represent technical replicates, whereas the color of each dot indicates the experimental origin (3 independent experiments; 4–60 technical replicates). Significance has been assessed by a two-tailed unpaired 2Student’s t test or two-tailed Mann-Whitney test when data were not normally distributed (∗p < 0.05, ∗∗∗∗p < 0.0001, ns, not significant).