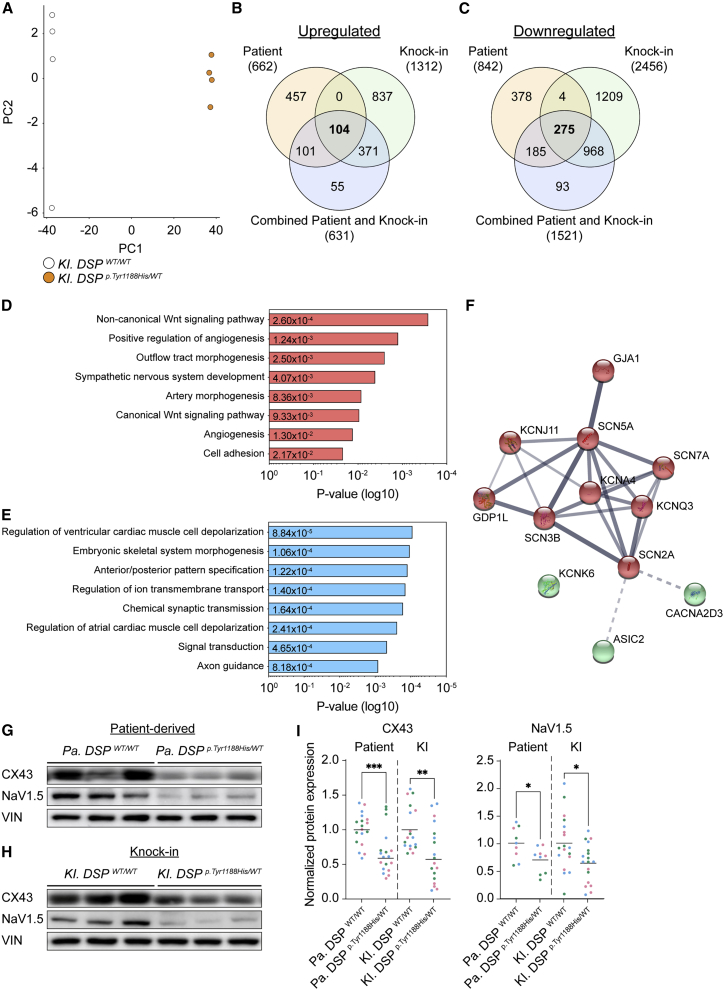
Figure 5.
Pa. DSPp.Tyr1188His/WT and KI. DSPp.Tyr1188His/WT cardiomyocytes display impaired ion-handling
(A) PCA reveals different gene expression profiles of KI. DSPWT/WT and KI. DSPp.Tyr1188His/WT cardiomyocytes.
(B–F) Combined analyses of the Pa. DSPp.Tyr1188His/WT (patient) and KI. DSPp.Tyr1188His/WT (knockin) mRNA sequencing datasets.
(B and C) Venn diagram showing the overlap of the up- (B; fold change (log2) >1) and downregulated (C; fold change (log2) <−1) genes.
(D and E) Gene Ontology analyses on the up- (D) and downregulated (E) genes overlapping between the two datasets.
(F) STRING: functional protein association network for the gene ontology terms “regulation of ventricular cardiac muscle cell depolarization” and “regulation of ion transmembrane transport.” Clustered based on kmeans. The thickness of each line indicates the level of confidence (data supported) and the colors to which cluster each component belongs.
(G–H) Representative immunoblots for CX43 and NaV1.5 in Pa. DSPp.Tyr1188His/WT (G) and KI. DSPp.Tyr1188His/WT (H) and isogenic control cardiomyocytes.
(I) Quantification of CX43 and NaV1.5 protein levels. Values normalized to VIN. Data are plotted as mean.
The dots in (I) represent technical replicates, whereas the color of each dot indicates the experimental origin (3 independent experiments; 4–6 technical replicates). Significance has been assessed by a two-tailed unpaired Student’s t test (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).