Figure 2.
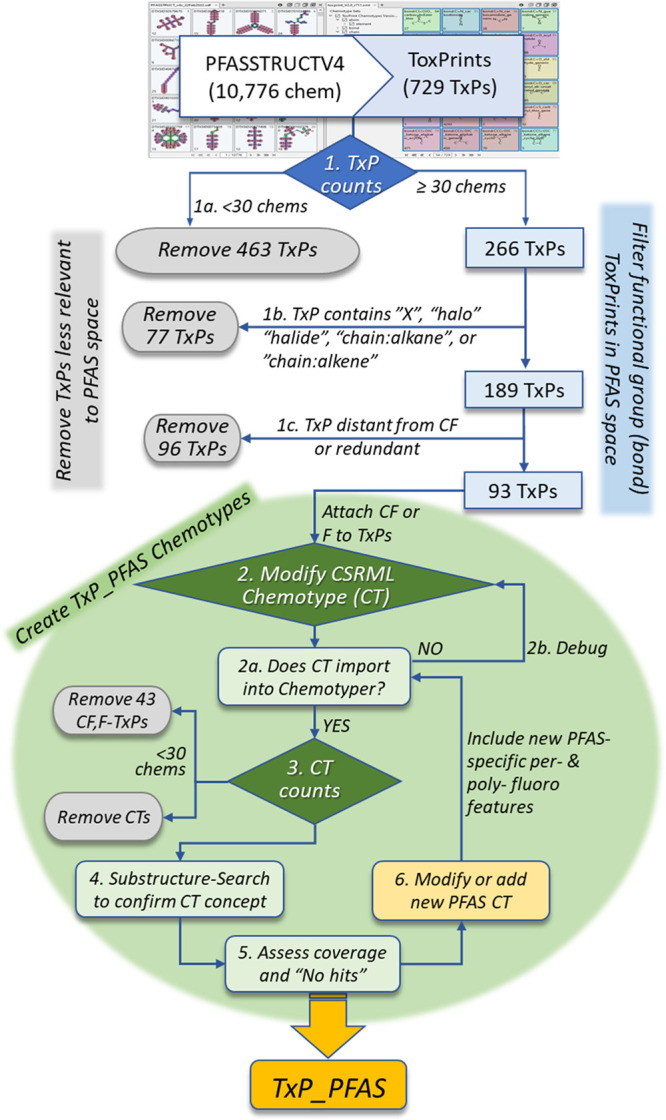
Workflow steps used to create the initial TxP_PFAS fingerprint set: Step 1. match 10,776 PFASSTRUCTV4 structures to 729 ToxPrints (TxPs) in the ChemoTyper, export the fingerprint file, generate total TxP counts; 1a. remove 463 TxPs, each in fewer than 30 chemicals; 1b. remove 77 TxPs whose name includes the terms “X”, “halogen”, “halide”, “chain:alkane” or “chain:alkene”; 1c. remove 96 TxPs found by manual inspection to be distant from the perfluoro portion of the structure, or redundant or closely related to another TxP; 2. for the remaining TxPs, either a CF or an F (depending on the feature) is added to the CSRML chemotype (CT) block; 2a. check to see that the CT imports into the ChemoTyper and the concept is properly visualized and conveyed; 2b. if not, manually inspect and debug the CT CSRML code and reimport; 3. import the CT into the ChemoTyper and generate total CT counts, removing 43 CTs in fewer than 30 chemicals from further consideration; 3a. assess how well the CT covers the feature space within PFASSTRUCTV4; 4. use substructure searching to independently validate the CT concept; 5. assess coverage of the individual CT and collection of TxP_PFAS CTs across the entire PFASSTRUCTV4 inventory and examine “NoHits” for possible missing features; 6. modify existing CT(s) or propose new CT(s) attached to a fluorine or fluoroalkyl moiety and introduce new PFAS-specific per- and polyfluoro features to capture capped (CF3) and noncapped (CF2) chains, partial hydrogenation, fluorotelomers, alternative halogenation (Cl, Br, I), branching, etc.
