Figure 4. METTL8 deletion reduces mitochondria protein expression and impairs radial glia neuronal stem cell maintenance in human forebrain organoids.
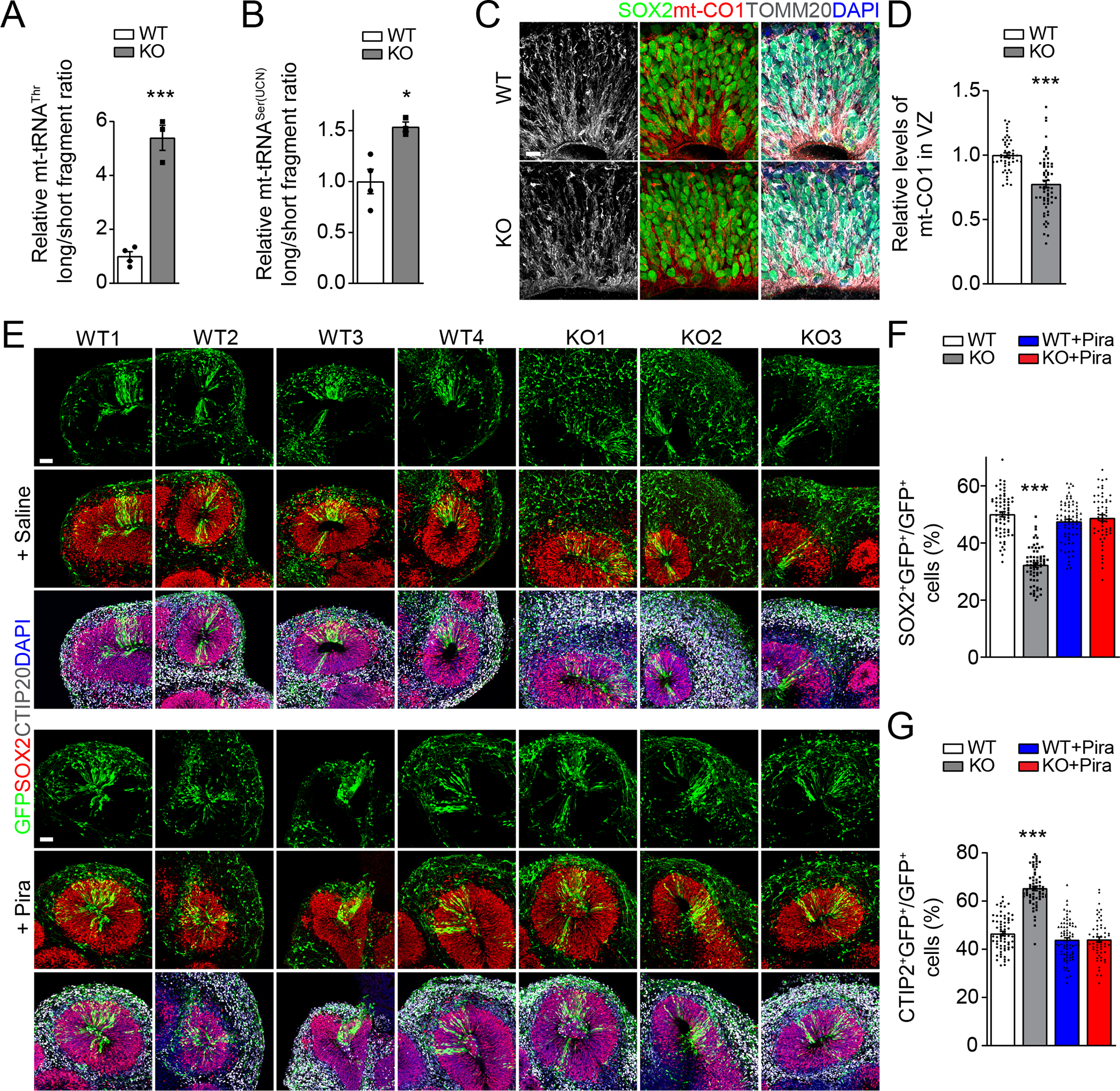
(A-B) Reduced m3C modification on mt-tRNAThr/Ser(UCN) in day 33 (D33) METTL8 KO organoids. Similar as in Figures 1C–D. Individual dots represent data from organoids derived from each iPSC line. Values represent mean ± SEM (n = 4/WT, 3/KO iPSC lines; *P < 0.05; ***P < 0.001; Student’s t-test).
(C-D) Sample immunostaining confocal images (C; Scale bar: 10 μm) and quantification of the relative intensity of mt-CO1 signal after normalization with the number of SOX2+ neural stem cells at the VZ regions of WT and KO D33 organoids (D). Individual dots represent data from each rosette of organoids derived from either WT or KO iPSC lines. Values represent mean ± SEM (n = 48 rosettes from 4 WT iPSC lines, 61 rosettes from 3 KO iPSC lines; ***P < 0.001; Student’s t-test).
(E-G) Sample immunostaining confocal images (E; Scale bars: 50 μm) for human organoids from each WT and KO iPSC line with or without treatment of piracetam (1 mM; from D34 to D56), injected with GFP-expressing retrovirus at D42 and analyzed 14 days later and quantification of percentages of SOX2+GFP+ neural stem cells (F) and CTIP2+GFP+ neurons (G) among all GFP+ cells in WT and KO organoids at D56. Values represent mean ± SEM (n = 68 sections from 4 WT iPSC lines, 64 sections from 3 KO iPSC lines, 76 sections from 4 WT iPSC lines with Pira treatment, 56 sections from 3 KO iPSC lines with Pira treatment; ***P < 0.001; One-way ANOVA).
See also Figure S4
