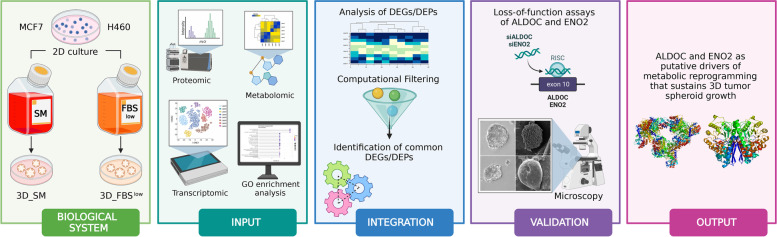
Fig. 1.
Workflow of the multi-omics integrative analysis. Biological System: H460 LUAD and MCF7 breast cancer cell lines were cultured in 2D and 3D conditions. 3D tumor spheroids were grown either in a nutrient-rich (sphere medium, SM) or in a nutrient-restricted (FBSlow) culture media. Input: a total of 6 samples (H460 2D, H460 3D_SM, H460 3D_ FBSlow, MCF7 2D, MCF7 3D_SM, MCF7 3D_ FBSlow) were characterized through transcriptomic, proteomic and metabolomic analyses; differentially expressed genes (DEGs) and differentially expressed proteins (DEPs) between 3D vs 2D samples were further analyzed by using Gene Ontology (GO). Integration: DEGs and DEPs were integrated to identify a common signature of DEGs and DEPs in all the 2D to 3D transitions. ALDOC and ENO2 were found up-regulated in all 3D vs 2D culture conditions. Validation: siRNA-mediated knock down of ALDOC and ENO2 were performed to functionally validate the effects of these two enzymes on 3D tumor spheroids growth. Output: ALDOC and ENO2 represent putative drivers of the metabolic reprogramming responsible for the sphere-forming ability of H460 and MCF7 cells