Figure 5.
Peptides encoded by sORFs3–15 aa have distinct interaction profiles
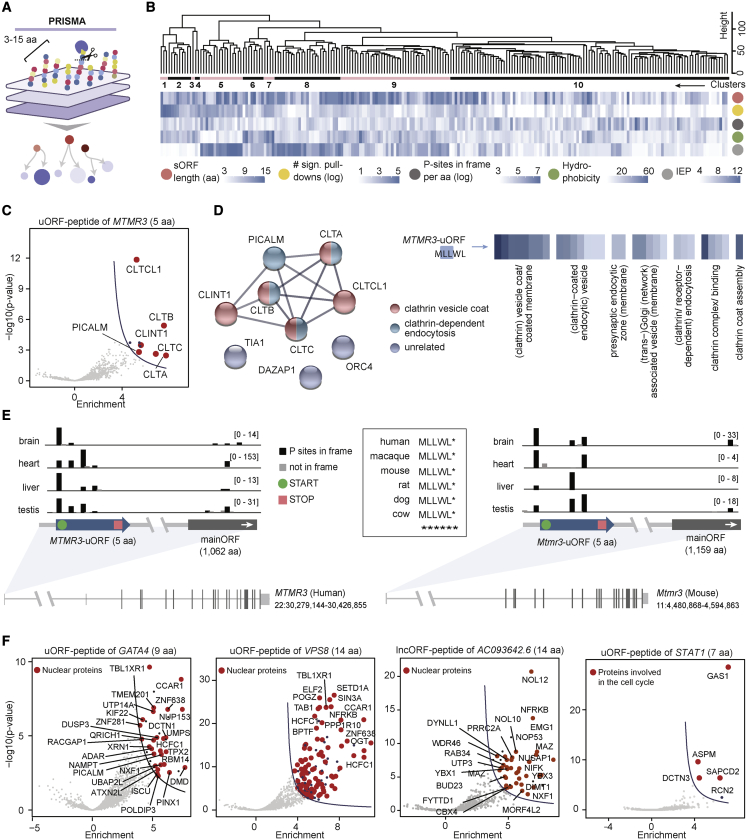
(A) Schematic of the PRISMA approach with all 221 sORF-encoded peptides3–15 aa.
(B) Hierarchical clustering of the enrichment values of all interacting proteins per peptide3–15 aa (STAR Methods). Factors potentially influencing the clustering (length, number of pull-downs per peptide3–15 aa [logarithmic scale], in-frame P-sites per aa [logarithmic scale], hydrophobicity, and isoelectric point) are depicted below the heatmap.
(C) Volcano plot of the peptide encoded by MTMR3-uORF. Proteins assigned to the GO term clathrin-dependent endocytosis (GO:0072583) as well as the clathrin-binding protein CLINT1 are highlighted in red.
(D) Left: string network of all significantly bound proteins of the MTMR3-uORF-peptide. Lines indicate confidence based on experiments, databases, and co-occurrence; high confidence (0.7). Right: MTMR3-uORF-peptide sequence (di-leucine motif highlighted) and GO enrichment analysis of its interactors.
(E) Genomic view and sequence alignment of the highly conserved MTMR3-uORF locus in four human (left) and four mouse (right) tissues.22,23
(F) Volcano plots summarizing the PRISMA results of the peptides3–15 aa translated from GATA4-uORF, VPS8-uORF, AC093642.6-lncORF, and STAT1-uORF.