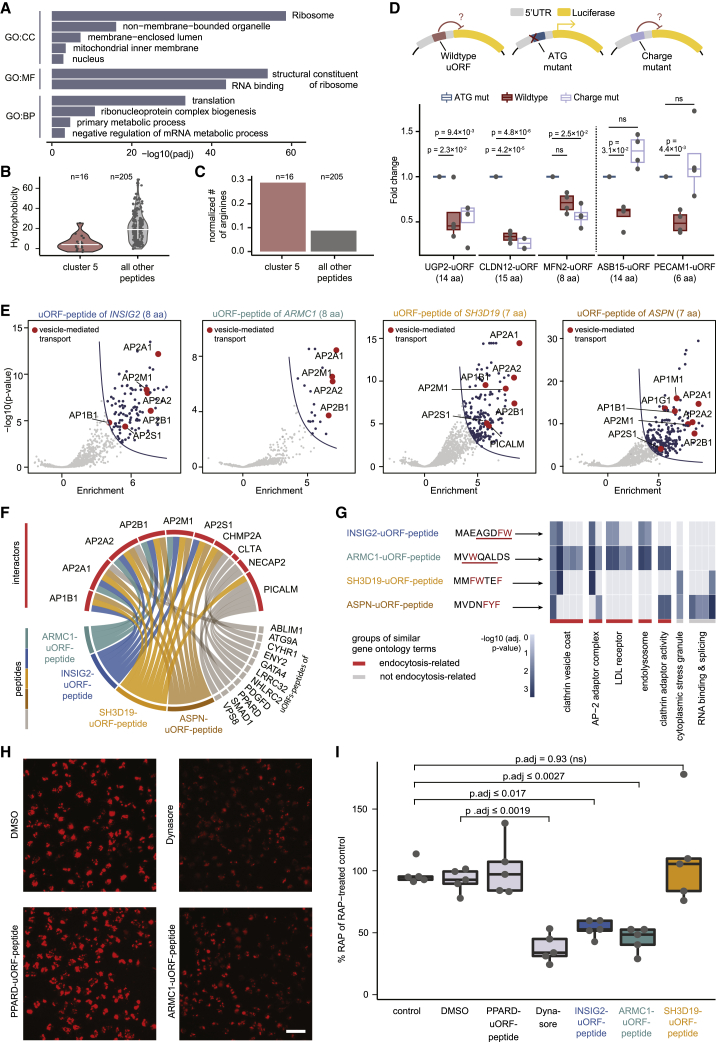
Figure 6.
Peptide interactomes can predict modulators of cellular functions
(A) GO enrichment analysis of all interacting proteins of 16 ribosome-binding peptides3–15 aa compared with all other peptides.
(B) Violin plot with hydrophobicity values of the 16 ribosome-binding peptides3–15 aa compared with all other peptides3–15 aa. Horizontal lines indicate the mean ± standard deviation.
(C) Number of arginines of the 16 ribosome-binding peptides3–15 aa compared with all other peptides3–15 aa, normalized to the total number of amino acids.
(D) Schematic and results of the luciferase reporter assay performed with five randomly selected ribosome-binding peptides3–15 aa. The significance was calculated using ANOVA and Tukey post hoc test.
(E) Volcano plots of four AP-binding peptides3–15 aa. Proteins assigned to the GO term vesicle-related transport (GO:0016192) are highlighted in red.
(F) Circos plot of all peptides3–15 aa that interact with endocytic proteins.
(G) Peptide sequences of the four AP-binding peptides3–15 aa (aromatic aa highlighted in red, di-hydrophobic motifs underlined) and GO enrichment analysis of their interactomes.
(H) Representative immunofluorescence images of fluorescently labeled RAP internalized by BN16 cells treated with DMSO, dynasore, PPARD- and ARMC1-uORF-peptide, respectively. Scale bar represents 200 μm.
(I) Results of the RAP endocytosis assay (five replicates per condition). Values were normalized to total protein content, and samples without RAP treatment were subtracted and then normalized to the treatment with RAP only (=100%). The PPARD-uORF-peptide, which did not bind APs, was included as a control (Figure S6J). The statistical significance was calculated using ANOVA and Tukey post hoc test.