Figure 3. Increased neuronal Tet2 in the adult hippocampus alters hydroxymethylation on genes related to synaptic processes and impairs memory.
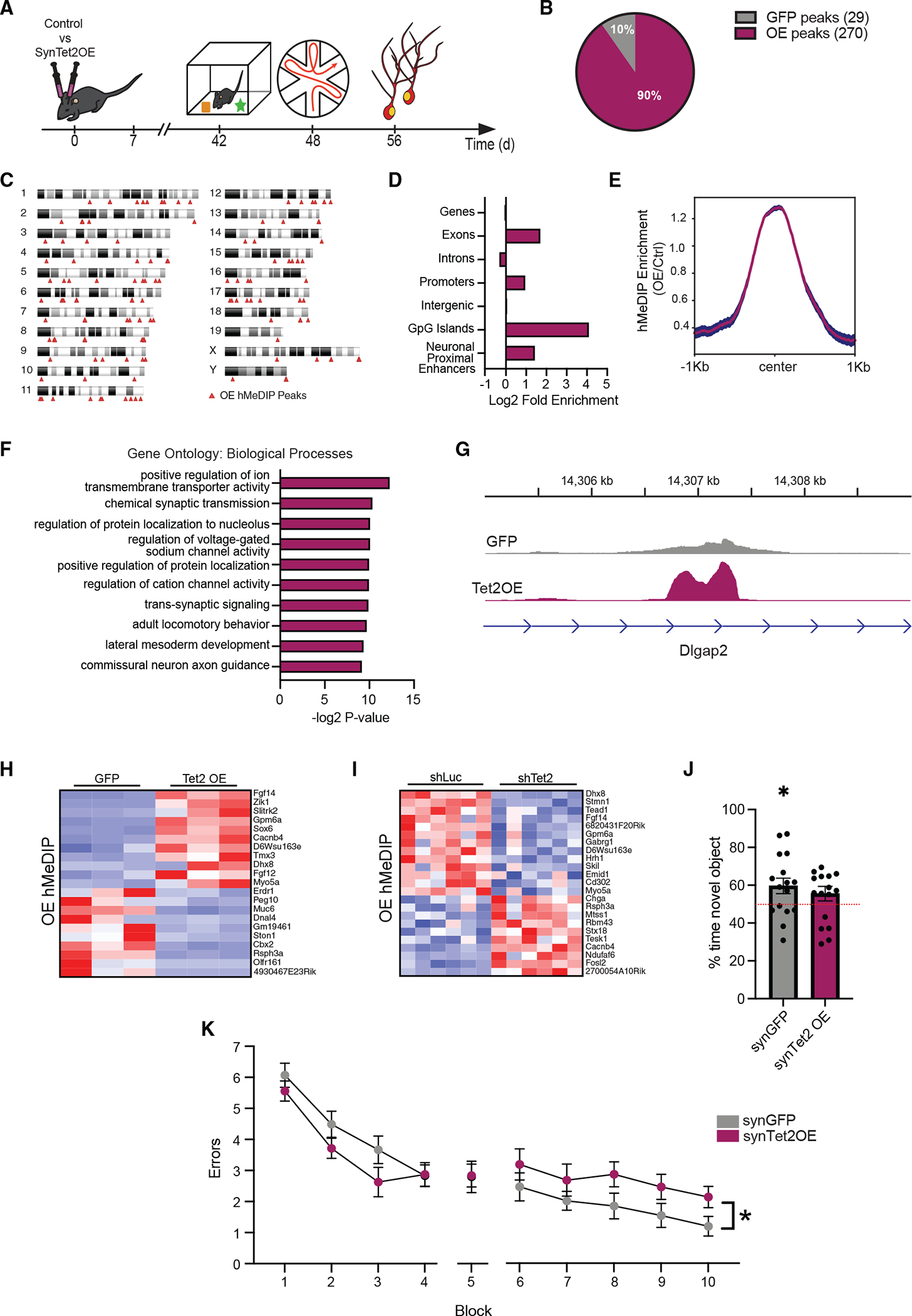
(A) Schematic of experimental paradigm. Adult (3–4 months old) wild-type mice were given bilateral stereotaxic injections of lentivirus (LV) encoding either Tet2 (Tet2 OE) or GFP control sequences driven by the neuron-specific Synapsin1 promoter into the hippocampus and subject to behavioral and molecular analysis 5 weeks later.
(B) Pie chart of 5hmC peaks gained (270 peaks) and lost (29 peaks) in Tet2 OE neurons compared with GFP control (q = 0.05).
(C) Ideogram of genomic location of hMeDIP peaks gained following neuronal Tet2 overexpression.
(D) Enrichment of gained hMeDIP peaks over genomic elements in Tet2 OE neurons.
(E) Metagene profile of gained hMeDIP peaks following neuronal Tet2 overexpression. Mean ± SEM.
(F) Gene Ontology analysis of biological processes on the 131 genes associated with the 270 gained hMeDIP peaks following neuronal Tet2 overexpression.
(G) Example gene track of the gained hMeDIP peak associated with Dlgap2 in Tet2 OE neurons.
(H) Heatmap of differentially expressed genes in the in vitro Tet2 OE RNA-seq dataset that have associated gained hMeDIP peaks in the in vivo Tet2 OE hMeDIP-seq dataset.
(I) Heatmap of differentially expressed genes in the in vitro shTet2 RNA-seq dataset that have associated gained hMeDIP peaks in the in vivo Tet2 OE hMeDIP-seq dataset.
(J) Quantification of percentage of interaction time during novel object recognition testing (n = 15–16 mice per group; t test, *p < 0.05).
(K) Quantification of the number of entry errors during radial arm water maze training and testing (n = 15–16 mice per group; repeated-measures ANOVA with Bonferroni post hoc correction, *p < 0.05).
Data are represented as mean ± SEM
