Figure 4. Loss of Tet2 in adult excitatory neurons alters hydroxymethylation on genes related to synaptic processes and enhances hippocampal-dependent memory.
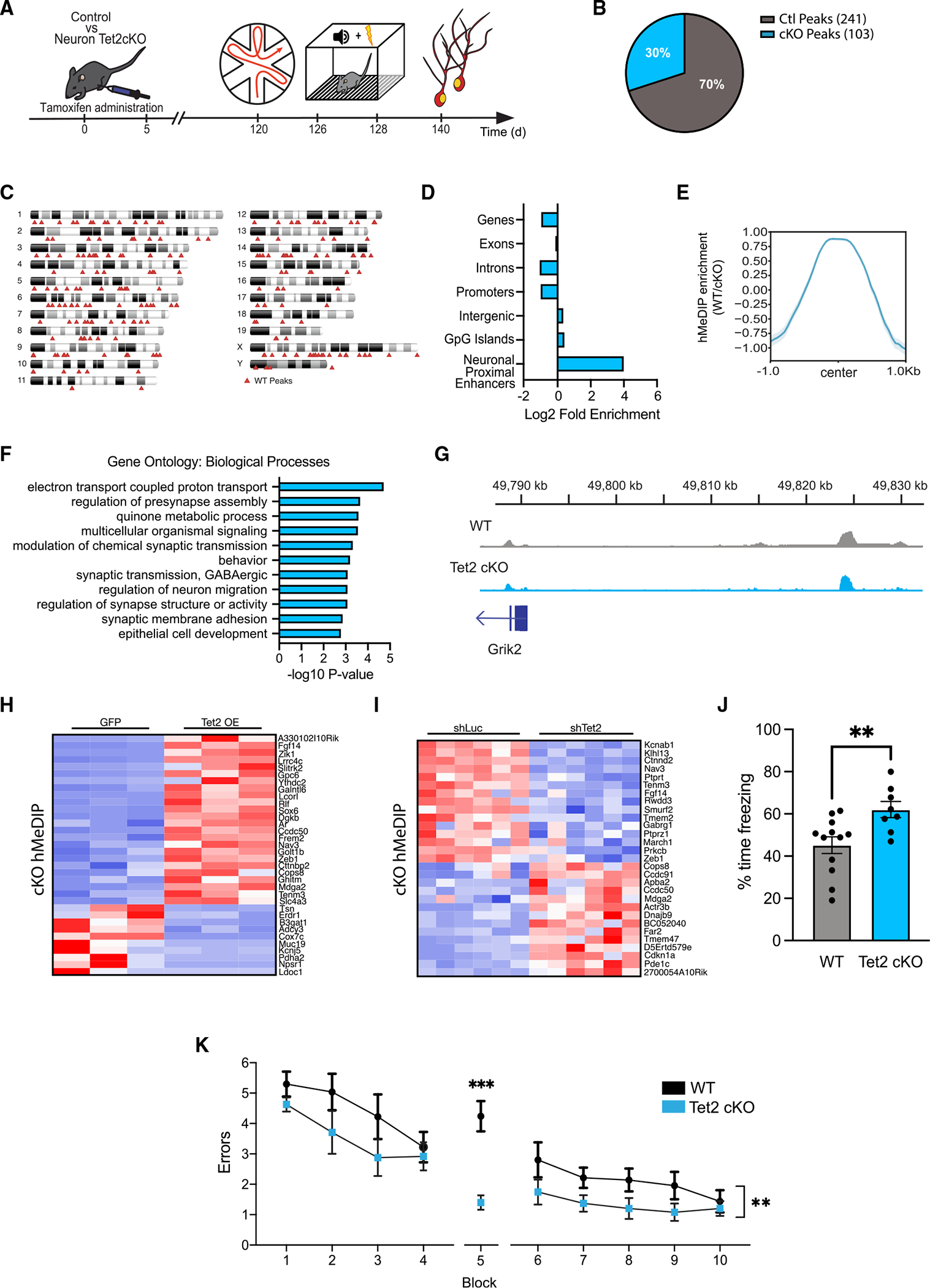
(A) Schematic of experimental paradigm. Adult (4 months old) CamK2aCre-ERT2;Tet2flox/flox excitatory neuron-specific knockout (Tet2 cKO) and littermate Tet2flox/flox control (wild-type [WT]) mice were administered tamoxifen and subjected to behavioral and molecular assays after 4 months.
(B) Pie chart of hMeDIP peaks gained (103 peaks) and lost (241 peaks) in Tet2 cKO neurons compared with control (q = 0.05).
(C) Ideogram of genomic location of hMeDIP peaks lost following loss of neuronal Tet2.
(D) Enrichment of lost hMeDIP peaks over genomic elements in Tet2 cKO neurons. (E) Metagene profile of lost hMeDIP peaks following loss of neuronal Tet2. Mean ± SEM.
(F) Gene Ontology analysis of biological processes on the 202 genes associated with the 241 lost hMeDIP peaks following loss of neuronal Tet2.
(G) Example gene track of the lost hMeDIP peak associated with Grik2 in Tet2 cKO neurons.
(H) Heatmap of differentially expressed genes in the in vitro Tet2 OE RNA-seq dataset that have associated lost hMeDIP peaks in the in vivo Tet2 cKO hMeDIP-seq dataset.
(I) Heatmap of differentially expressed genes in the in vitro shTet2 RNA-Seq dataset that have associated lost hMeDIP peaks in the in vivo Tet2 cKO hMeDIP-seq dataset.
(J) Quantification of the percentage of freezing 24 h after contextual fear conditioning training is shown (n = 8–12 mice per group; t test, **p < 0.01).
(K) Quantification of the number of entry errors during radial arm water maze training and testing (n = 8–12 mice per group; repeated-measures ANOVA with Bonferroni post hoc correction, **p < 0.01, ***p < 0.001).
Data are represented as mean ± SEM
