Extended Data Fig. 1. Filament formation in PRPS1.
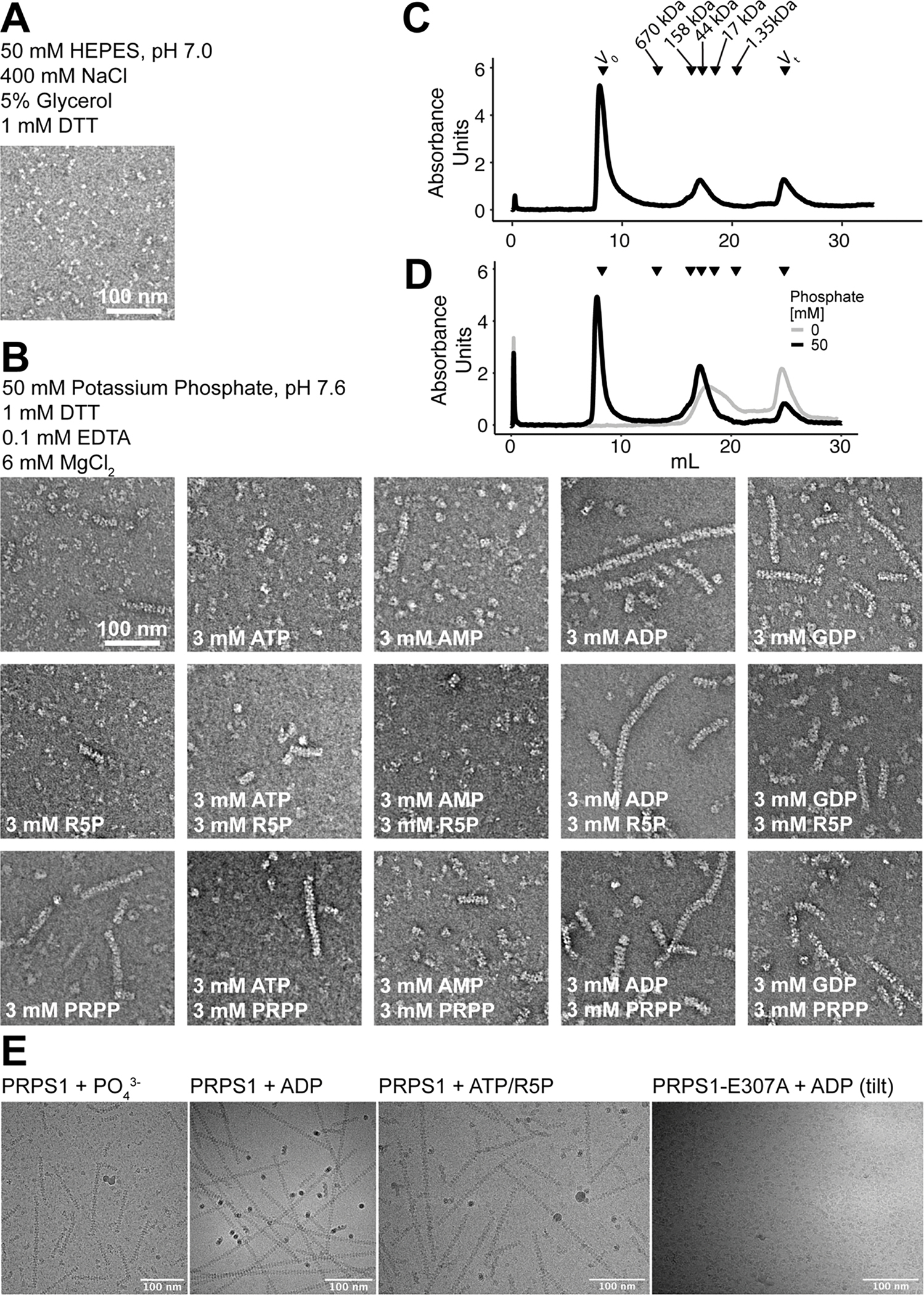
A. Section of negative stain EM of purified PRPS protein in a HEPES/Salt buffer used for purification. B. Panel of negative stain EM sections of PRPS1 in phosphate buffer in the presence of the indicated ligands. C. Elution profile from a size exclusion column (Superose 6) of PRPS1 in 50 mM phosphate buffer, pH 7.6. D. Elution profile from a size exclusion column (Superose 6) of PRPS1 in 100 mM KCl, 50 mM HEPES, pH 7.6 in the presence (black) or absence (grey) of 50 mM potassium phosphate, pH 7.6. PRPS1: monomer, 35 kDa; dimer, 70 kDa; hexamer, 210 kDa; filament, ≧ 420 kDa. E. Motion-corrected and summed cryo electron micrographs, Gaussian blurred and contrast adjusted for visualization, from four datasets presented in this manuscript, representing the cameras and tilts used in data collection. Microscopes, cameras, and stage tilts are listed in Data Table 1.
