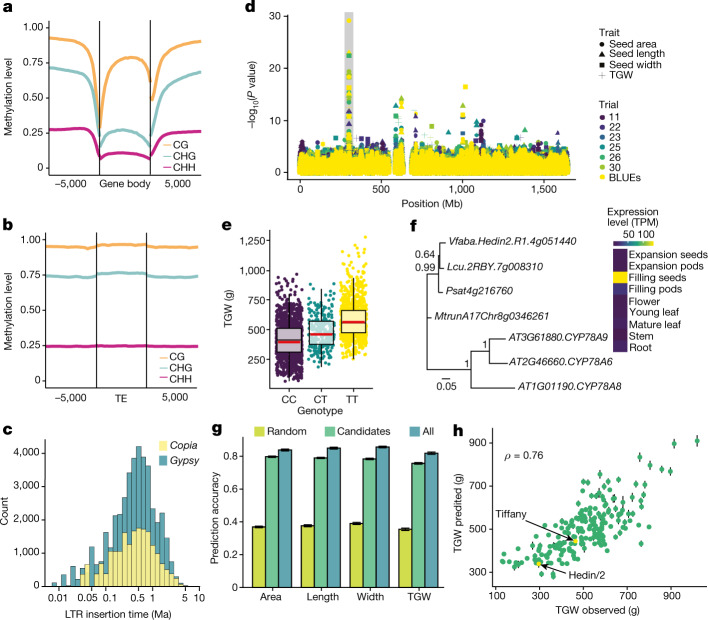
Fig. 3. TE methylation and seed size genetics.
a, Global distribution of DNA methylation levels at protein-coding genes including a 5-kb region upstream of the transcription start site and downstream of the transcription end site. b, DNA methylation patterns for TEs and their 5-kb flanking regions. c, Distribution of Copia (RLC) and Gypsy (RLG) retrotransposons based on insertion time. d, Combined Manhattan plot of GWAS analysis of seed area, seed width, seed length and thousand grain weight (TGW) for chromosome 4. BLUE, best linear unbiased estimator. e, Effect plot for TGW for the SNP marker within Vfaba.Hedin2.R1.4g051440 at position 299,823,118 on chromosome 4 (highlighted by a grey bar in part d). n = 2,499 data points distributed over six trials. In the box plots, the horizontal black central line represents the median, the red line indicates the mean, the box ranges from the first to third quartile, and the vertical black lines extend to the smallest or largest point within the 1.5× interquartile range. f, Phylogenetic tree showing the relationships between Vfaba.Hedin2.R1.4g051440 and its orthologues in pea, lentil, Medicago and Arabidopsis and its expression levels in nine diverse tissues of Hedin/2 in transcripts per million (TPM). The branch lengths are measured in the number of substitutions per site. Numbers next to branches indicate bootstrapping support. g, Genomic best linear unbiased prediction accuracies for seed traits using 15 randomly chosen SNPs (random), the 15 seed-size-associated markers (candidates) or all markers (all). h, Mean genomic best linear unbiased prediction of TGW using the 15 seed-size-associated SNPs plotted against the mean observed values. The Pearson correlation coefficient is indicated. Error bars indicate the standard deviation of a fivefold genomic best linear unbiased prediction cross-validation scheme with predicted values computed from ten replicate runs (g,h; see Methods).