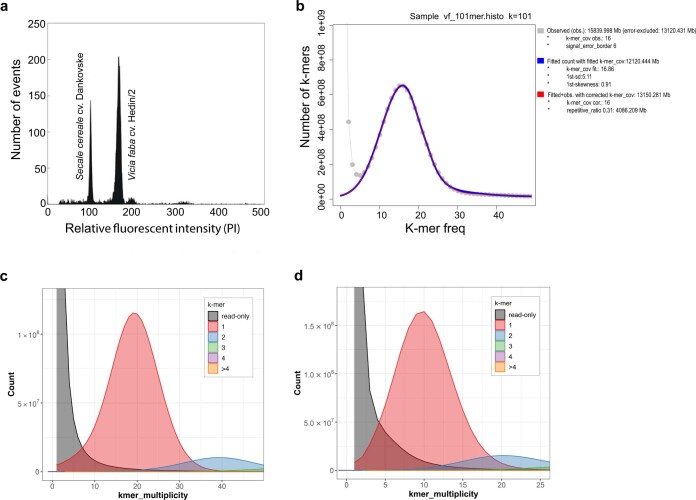
Extended Data Fig. 1. K-mer and flow-cytometry based genome size estimation.
a, Estimation of nuclear genome size of V. faba Hedin/2 using flow cytometry. The histogram of relative nuclear DNA content was obtained after flow cytometric analysis of propidium-iodide stained nuclei of V. faba ‘Hedin/2’ and Secale cereale cv. Dankovske (2C = 16.19 pg), which served as the internal reference standard. The low level threshold was set to channel 20 on a 512-channel scale histogram of relative fluorescence intensity; all remaining fluorescence events were recorded. 2C nuclear DNA content of V. faba Hedin/2 was estimated at 26.36 pg (± 0.26 s.d.). b, k-mer based estimation of genome size using Hedin/2 raw HiFi sequencing data. c, k-mer spectrum plot produced by Merqury for the Hedin/2 genome assembly. K-mer multiplicity refers to the number of times a k-mer is found in sequencing reads. Colour corresponds to the number of times a k-mer is found in the assembly. d, k-mer spectrum plot produced by Merqury for the Tiffany genome assembly.