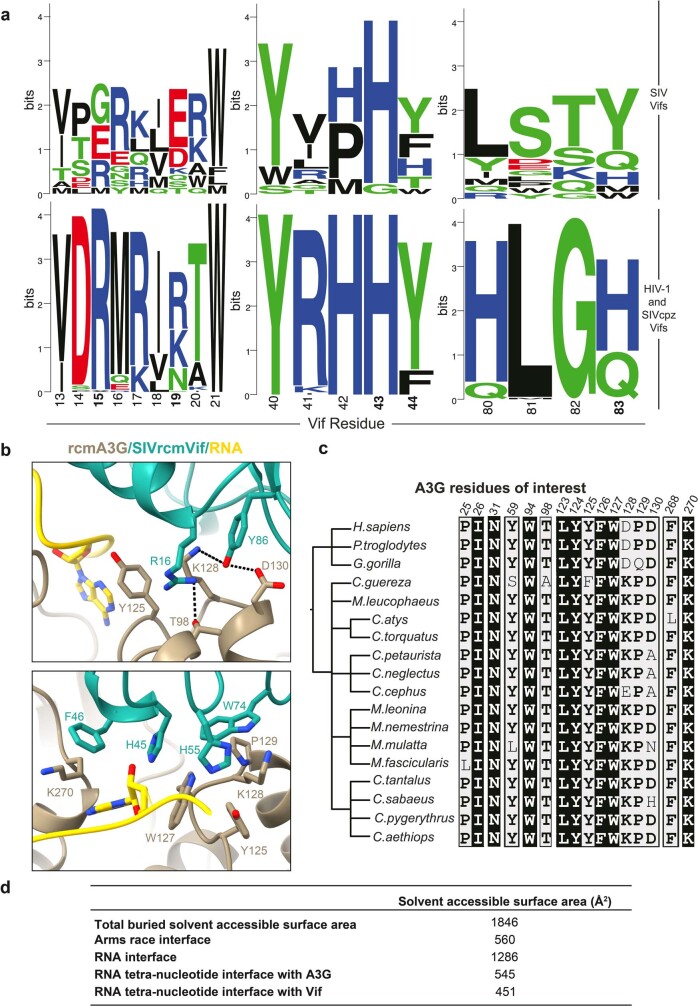
Extended Data Fig. 6. Characterization of arms race interface.
a, Logo plots showing sequence variation of Vif residue that interacts with A3G for HIV-1 and SIVcpz (bottom) and other SIVs (top). Data is similar to that of Fig. 2c except that neighboring residues are also shown for context. b, A comparative model of rcmA3G–SIVrcm Vif–CBFβ was built with the hA3G-VCBC monomer structure as a template. Dashed lines represent the hydrogen bond network involved in the arms race interface (top). Residue K128 of rcmA3G interacts with Y86 of SIVrcm Vif, the primary determinant of Vif adaption to counteract hominid A3G50. Residue F46 and W74 of SIVrcm Vif previously reported to be critical for rcmA3G neutralization engage in extensive hydrophobic interactions with rcmA3G in the model (bottom). Note amino acids 16 and 86 of SIVrcm Vif correspond to amino acids 15 and 83, respectively, in HIV-1 Vif. c, Sequence alignment of A3G residues that contact RNA or Vif from Old World Monkeys and hominids. Fully conserved residues are highlighted with white text on black background. d, Buried solvent accessible surface area for A3G–RNA–VCBC monomer structure.