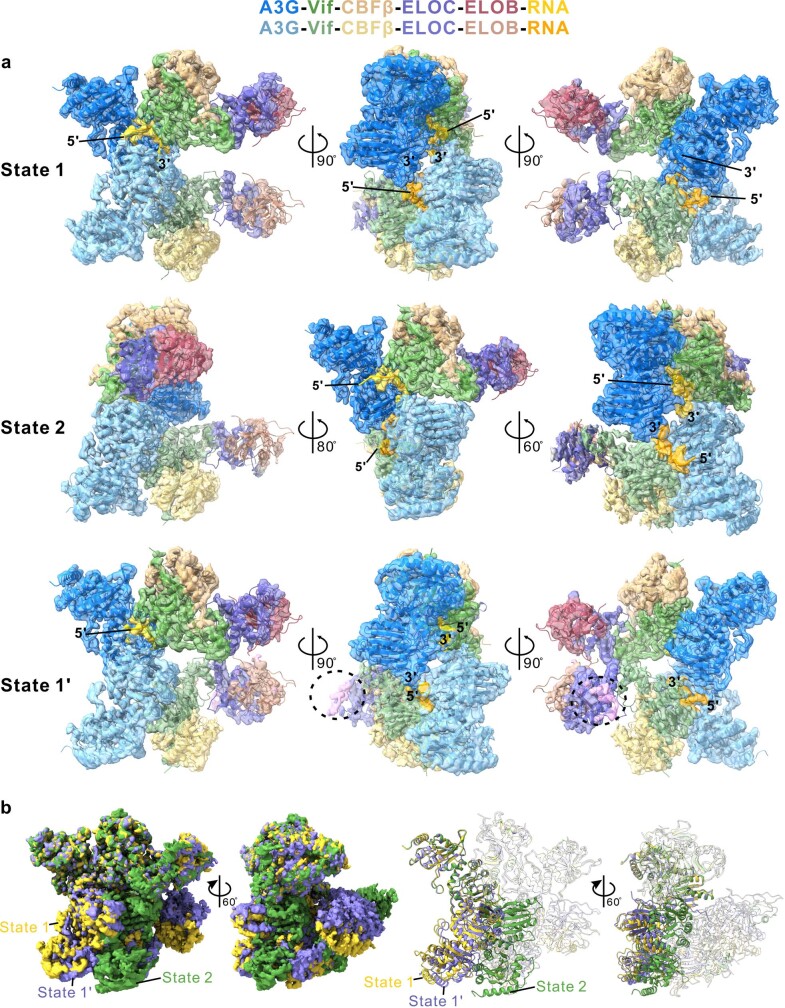
Extended Data Fig. 7. A3G-RNA-VCBC forms multiple discrete dimeric configurations.
a, Cryo-EM maps for State 1 (top), State 2 (middle), and State 1′ (bottom) colored by subunit, showing three dimeric configurations of the A3G–RNA–VCBC complex (Extended Data Fig. 2b). Densities for single copies of RNA in State 1 and State 2 are clear for 8 and 9 nucleotides, respectively. In contrast, only 5 nucleotides were fit in the density map for State 1′ due to the moderate resolution at both ends of RNA. The 5′ and 3′ ends of single-stranded RNA are indicated. Extra weak density (pink) near ELOC corresponds to the expected position of CUL5N (denoted by black dashed circle) is observed for State 1′. State 1 and State 2 differ by a rigid-body motion with the second A3G–VCBC protomer rotated by 66° and translated by 28 Å relative to one another (Supplementary Video 1). b, A comparison of cryo-EM maps for State 1 (yellow), State 1′ (purple), and State 2 (green) in two orientations (left). Corresponding view of the structures for State 1, State 1′, and State 2, which are aligned by superposing A3G monomer (right). A3G is shown in solid ribbon while other proteins are in transparent for clarity. State 1′ has a dimeric configuration much similar to State 1, which are related by a 9° rotation and 4 Å translation. See Supplementary Discussion for the details.