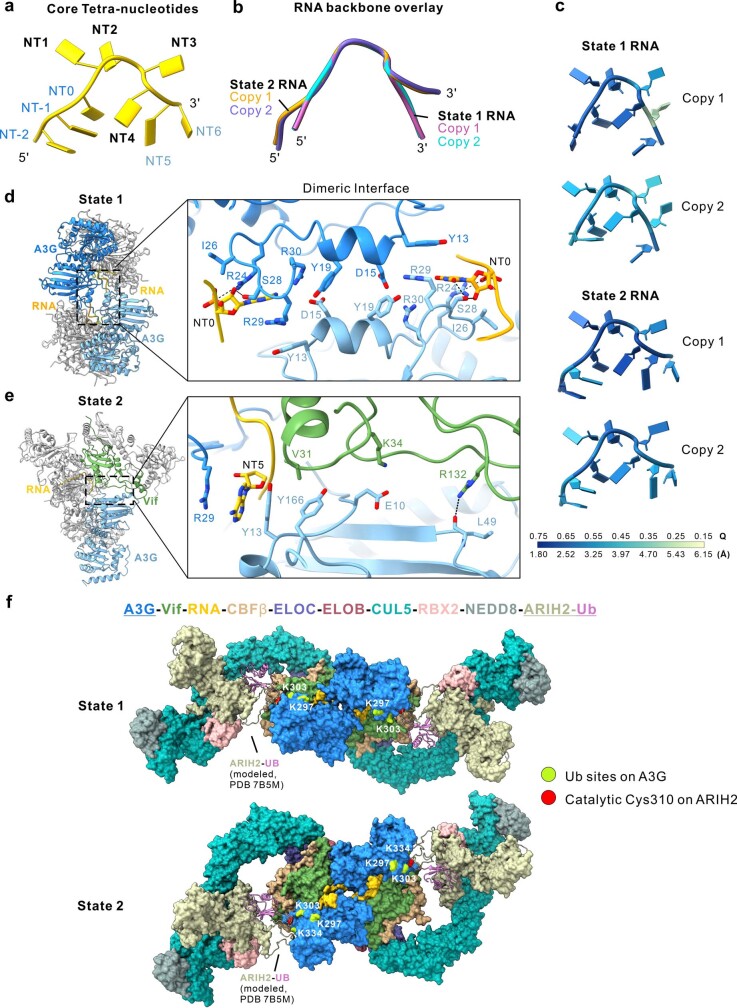
Extended Data Fig. 8. Dimeric interfaces in the different configurations of the A3G-RNA-VCBC complex.
a, Schematic illustrating the annotation of RNA nucleotides used in this study. NT1-4 denotes the core tetra-nucleotides buried in the groove formed by A3G and Vif in the monomer; NT0 and NT5 are located near the dimeric interface for State 1 and State 2, respectively. b, Overlay of two copies of RNA backbone for State 1 and State 2. c, RNA models colored by per nucleotide Q-score87. The corresponding resolution estimated from each Q-score is indicated at the bottom of colored bar. The Q-score for State 1 and State 2 EM map is 0.54 and 0.56, respectively. d–e, Structure overview (left) and close-up (right) of the dimeric interface formed by (d) A3G and A3G for State 1 and (e) A3G and Vif for State 2. The buried solvent accessible surface area calculated by Chimera X92 is ~ 350 and ~ 480 Å2, respectively. Note that the RNA interface is in close proximity to the dimeric interface, suggesting RNA aids or dominates dimerization for both State 1 and State 2. See Supplementary Discussion for the details. f, Model of dimeric A3G-RNA-VCBC in complex with CUL5/RBX2 E3 ligase bound to ubiquitin-loaded ARIH2 (ARIH2~UB) for State 1 (top) and State 2 (bottom). States 1 and States 2 of A3G-RNA-VCBC are compatible with CUL5/RBX2 binding and ubiquitin transfer. The model was built by overlaying A3G-RNA-VCBC dimer structures determined in this study with published structure VCBC-CUL5NTD (PDB code 4N9F), neddylated CUL5CTD–RBX2–ARIH2 (PDB code 7ONI), and a comparative model of ARIH2~UB (built based on a partial structure containing CUL1-RBX1-ARIH1-UB; PDB code 7B5M). Ubiquitin sites (K297, K303, and K334) on A3G are colored in lime, and catalytic Cys 310 on ARIH2 colored in red. See Supplementary Discussion for the details.