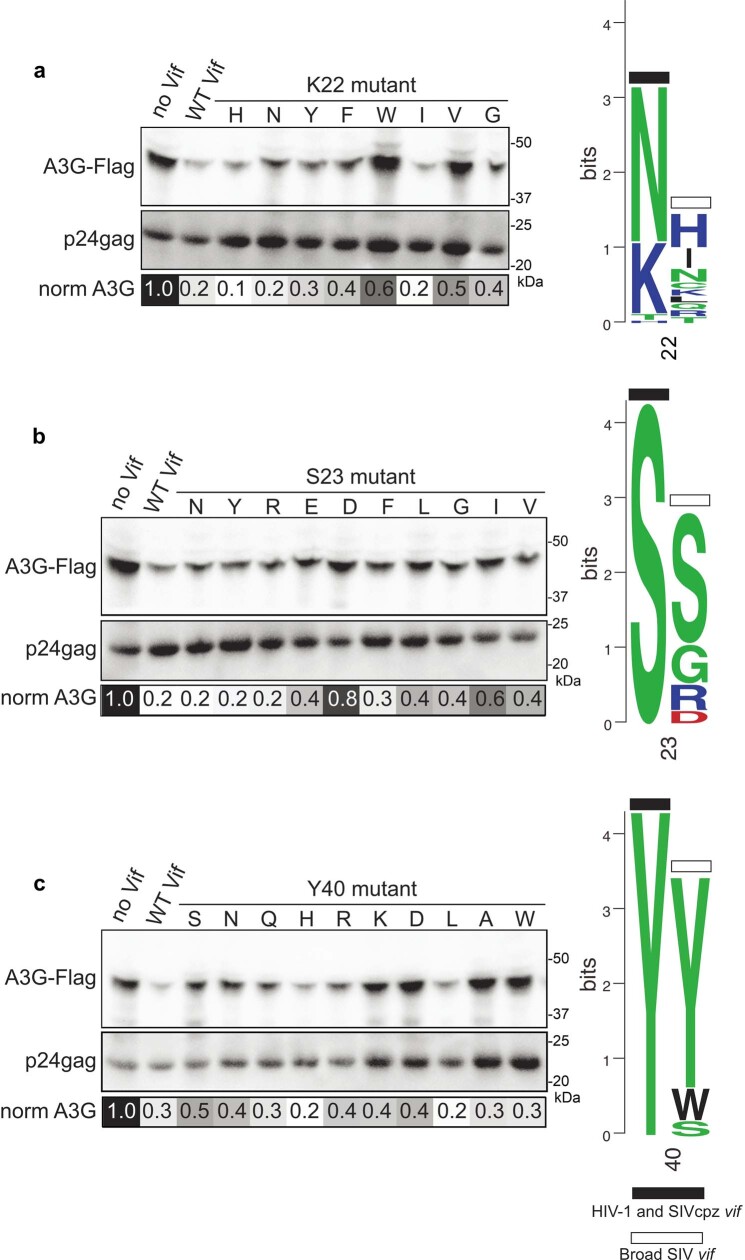
Extended Data Fig. 5. Functional and evolutionary assessment of Vif residues involved in RNA binding.
Left: Amino acid mutants at Vif residue a, K22, b, S23, and c, Y40 were assessed for their ability to prevent packaging of A3G into virions. Top Western blot in each panel shows the virion incorporation of A3G, while the bottom Western blot in each panel shows the amount of virus (p24gag) in the corresponding virion preparation. Below each panel is a greyscale heatmap of the relative A3G incorporation normalized to p24gag based on two replicate transfections (with the exception of S23Y and S23D) with the amount of A3G in the “No Vif” control set to 1.0 (darkest color). Controls were run on the same gel as the samples. For source data, see Supplementary Fig. 1a. Right of each panel: Logo plot of amino acids found in the consensus of all HIV-1 clades as well as SIVcpz (black bar), and all other SIV strains using equal distribution of each SIV (white bar).