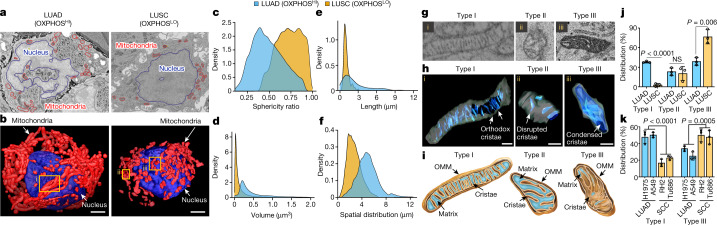
Fig. 3. Structural and spatial analysis of mitochondrial networks in SBEM-imaged NSCLC tumour volumes.
a, Representative 2D SBEM images of an OXPHOSHI LUAD cell and an OXPHOSLO LUSC cell. b, 3D reconstruction of nucleus (blue) and mitochondrial (red) networks segmented from NSCLC cells in a. Yellow boxes show elongated mitochondria (i) and fragmented mitochondria (ii,iii). Scale bars, 3 μm. c–f, Density plots measuring mitochondrial sphericity (c), volume (d), length (e) and spatial distribution relative to nucleus surface (f) for OXPHOSHI LUAD cells (n > 50,000 mitochondria) and OXPHOSLO LUSC cells (n > 22,000 mitochondria). g,h, SBEM images and 3D reconstruction of representative type I (i), II (ii) and III (iii) crista structures identified in OXPHOSHI LUAD cells and OXPHOSLO LUSC cells. Scale bars, 500 nm (h). i, Illustration of type I, II and III crista structures. Outer mitochondrial membrane (OMM), matrix and inner mitochondrial membrane (cristae) are indicated. j, Percentage of mitochondrial type I, II and III crista distribution in OXPHOSHI LUAD cells (n = 3 biological replicates, n > 1,200 mitochondria) and OXPHOSLO LUSC cells (n = 3 biological replicates, n > 750 mitochondria). Data are mean ± s.e.m. Unpaired two-tailed t-test. k, Percentage of type I and III crista distribution in human LUAD (H1975, A549) and SCC (RH2, Tu686) cells. Data are mean ± s.e.m. (n = 3 biological replicates, n > 2,000 mitochondria). Unpaired two-tailed t-test.