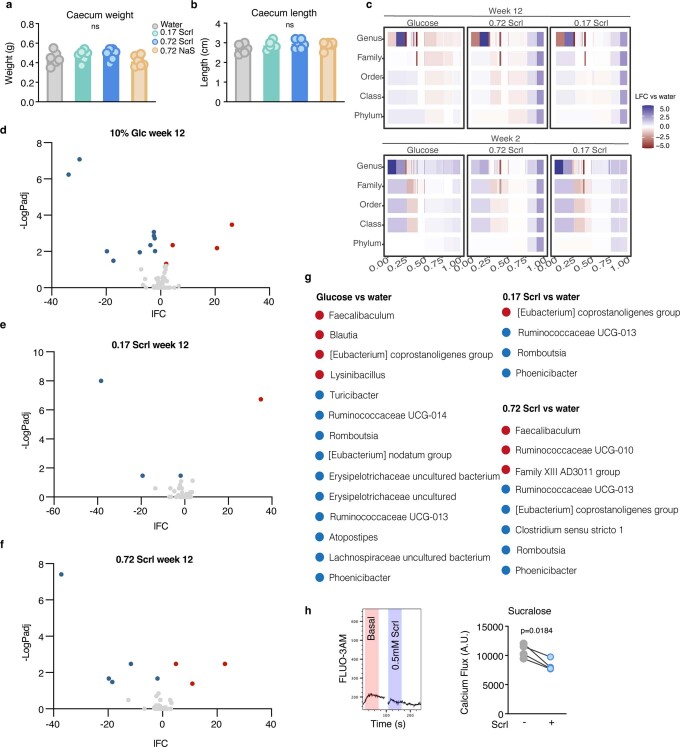
Extended Data Fig. 4. Microbiome analysis of mice treated with sweeteners and sucralose effect on calcium flux.
(a–b) C57BL/6J mice fed water, 0.17 mg ml−1 Scrl, 0.72 mg ml−1 Scrl or 0.72 mg ml−1 NaS for 12 weeks. Each dot represents an individual mouse. N = 6 per treatment. a) Caecum weight. b) Caecum length. (c–g) Gut microbiome analysis of mice fed for 2 and 12 weeks with water), 0.17 mg ml−1 Scrl, 0.72 mg ml−1 Scrl or 10% w/v glucose. N = 5 per treatment. c) Heatmaps showing alteration in abundance data in mice exposed to different drinking solutions as indicated compared to time-matched samples obtained from water-fed mice. Top graphs: samples collected after 12 weeks exposure; lower graphs samples collected after 2 weeks exposure. The variation of abundance is shown at Phylum, Class, Order, Family and Genus level. (d–f) Volcano plots of regulated genera in samples collected after 12 weeks of treatment. Water-treated animals were used as control. The fold changes were estimated using a negative binomial model from the DESeq2 package in R using its default settings for accounting for different library sizes between samples. A generalized linear model accounted for batch along with the interaction of treatment and time, to provide estimates of time effects within treatment (and vice versa). P-values were calculated using a Wald test and then adjusted using the Benjamini-Hochberg method to control for false discovery rate. Comparisons: d) 10% glucose vs water, e) 0.17 mg ml−1 Scrl vs water and f) 0.72 mg ml−1 Scrl vs water. Genera with a logarithmic fold change > 0.6 and an adjusted P-value < 0.05 are considered upregulated; Genera with a logarithmic fold change < 0.6 and an adjusted P-value < 0.05 are considered downregulated. g) List of upregulated (red) or downregulated (blue) genera in the different comparison as indicated in (d, e, f). h) Representative kinetics diagram (left) of FLUO-3AM calcium flux in response to 0.5 mM Scrl and corresponding quantification (right) of FLUO-3AM intensity in Jurkat T cells. N = 4 technical replicates. Data are representative of 3 independent experiments. The means represents ± s.e.m. for biological replicates (a, b), one-way ANOVA with Dunnet’s multiple comparison test (a, b), and 2-tailed paired Student’s t test (h) were used for statistical analysis.