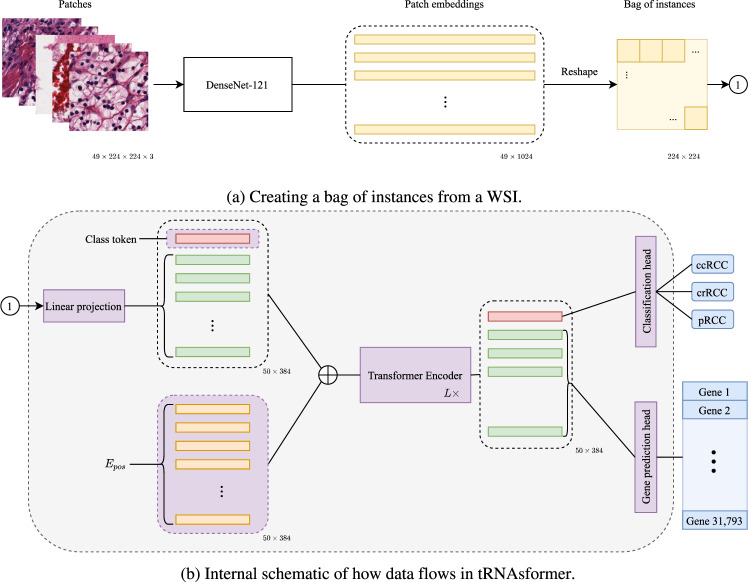
Fig. 1. A diagram showing how tRNAsformer works.
a 49 tiles of size 224 × 224 × 3 selected from 49 spatial clusters in a WSI are embedded with a DenseNet-121. The outcome is a matrix of size 49 × 1024 as DenseNet-121 has 1024 deep features after the last pooling. Then the matrix is reshaped and rearranged to 224 × 224 matrix in which each 32 × 32 block corresponds to a tile embedding 1 × 1024. b Applying a 2D convolution with kernel 32, stride 32, and 384 kernels, each 32 × 32 block has linearly mapped a vector of 384 dimensional. Next, a class token is concatenated with the rest of the tile embeddings, and Epos is added to the matrix before entering L Encoder layers. The first row of the outcome, which is associated with the class token, is fed to the classification head. The rest of the internal embeddings that are associated with all tile embeddings are passed to the gene prediction head. All parts with learnable variables are shown in purple.