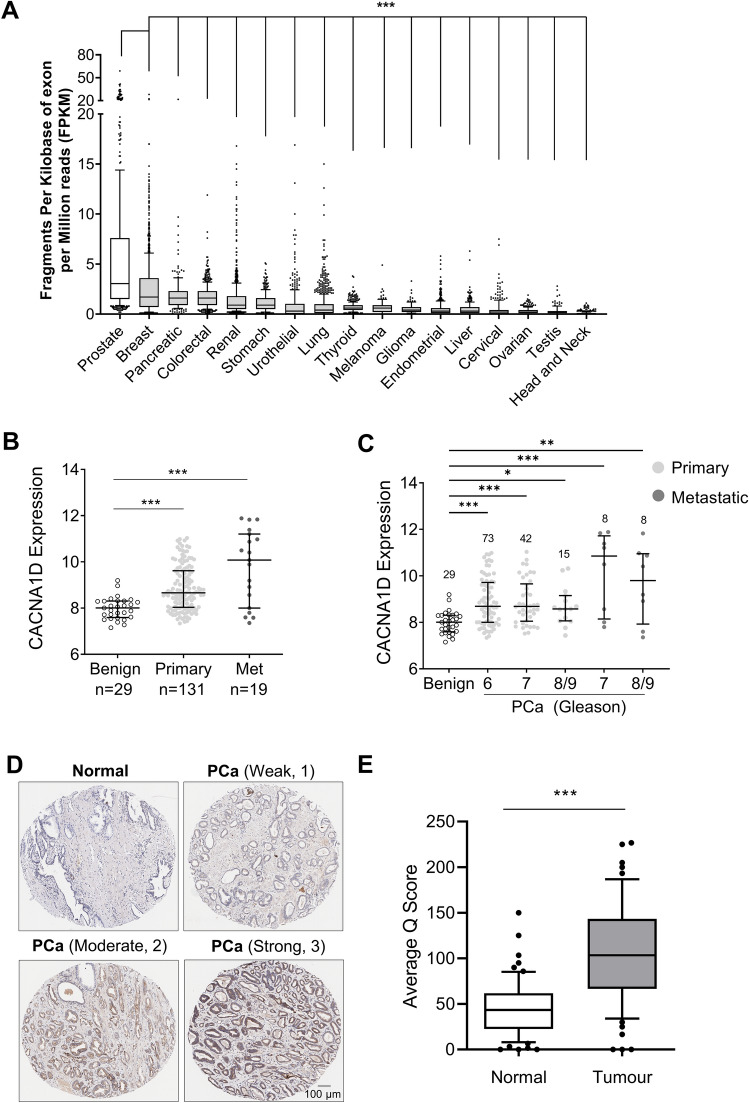
Figure 1.
Profiling CACNA1D and CaV1.3 expression in clinical PCa datasets and tissue microarrays. (A) Human CACNA1D transcript expression was determined by RNA-seq analysis from 7932 TCGA samples from 17 different cancer tissues. Data was obtained from The Human Protein Atlas (www.proteinatlas.org/) ENSG00000157388-CACNA1D/pathology (v21.1.proteinatlas.org), is presented as box-and-whisker plots (median, 10–90 percentile) and was analysed with Kruskal–Wallis, Dunn’s multiple comparison test p < 0.001 (***). (B) Scatter plot showing CACNA1D expression in primary (n = 131) PCa and metastatic PCa (n = 19) compared to adjacent benign prostate tissue (n = 29). Data was derived from GEO dataset, GSE21032 (GPL 5188) conducted by Memorial Sloan-Kettering Cancer Centre (MSKCC). Data is expressed as median and inter-quartile range and was analysed using Kruskal–Wallis, Dunn’s multiple comparison test, p < 0.001 (***). C. CACNA1D expression was compared by Gleason grade (6, 7 and 8–9) between primary and metastatic PCa samples. Data was derived from GEO dataset, GSE21032 (GPL 5188, MSKCC) and analysed using Kruskal–Wallis, Dunn’s multiple comparison test. Data is expressed as median and inter-quartile range where significance is indicated as p < 0.05 (*), p < 0.01 (**) and p < 0.001 (***). D. Examples of normal/benign and prostate tumour tissue cores from an in house tumour PCa microarray, stained with anti-CaV1.3 (HPA020215, Sigma Aldrich). Tissues were assessed by a Q scoring system. Staining intensity was scored as weak (1), moderate (2) and strong (3) and was multiplied by the percentage of tissue encompassed by the designated staining. (E) Q scores (median, 10–90 percentile) showing significant CaV1.3 overexpression in tumour vs normal/benign tissues (N = 67, Mann–Whitney, unpaired, p < 0.001 (***)).