Highlights
-
•
Non coding RNA and coding RNA date were integrated and analyzed.
-
•
Plasma exosome-derived circGAPVD1 could be a potential diagnostic biomarker for colorectal cancer (CRC) .
-
•
The ceRNA network of circRNA,miRNA and mRNA was constructed.
-
•
This study provides new data for the early diagnosis of colorectal cancer with non-coding RNA.
Keywords: Colorectal cancer, Exosomes, Circular RNA, Diagnosis, Biomarkers, ceRNA network
Abstract
Background
Although circular RNAs (circRNAs) have recently garnered interest as disease markers, they have been relatively poorly studied as a biomarker in colorectal cancer (CRC). In this study, we aimed to screen the exosome-derived circRNAs in CRC and explore their potential as diagnostic and prognostic biomarkers of CRC
Methods
Exosomes were extracted from the plasma using a kit and validated by immunoblotting, transmission electron microscopy, and particle size analysis. The microarray datasets were employed to identify differentially-expressed circRNAs from plasma exosomes. Real-time quantitative reverse transcription PCR (RT-qPCR) verified the results of the microarray analysis, and Receiver operating characteristic (ROC) curve revealed the diagnostic ability of a single circRNA. The Starbase combined with microT, miRmap, and RNA22 were used to establish a circRNA-miRNA-mRNA network. Gene ontology, Kyoto Encyclopedia of Genes, Genomes pathway enrichment analysis, and Gene Set Enrichment Analysis were applied to determine potential functions of the identified mRNAs
Results
Comparing the microarray of plasma exosome-derived circRNAs and the microarray downloaded from the GEO database, 15 candidate circRNAs with up-regulated expression were identified. RT-qPCR verified that hsa_circ_0003270 (circGAPVD1) was upregulated in CRC plasma exosomes. ROC analysis showed that circGAPVD1 in plasma exosomes has potential diagnostic value for CRC. The sensitivity and specificity of circGAPVD1 in the diagnosis of CRC were found to be 75.64 and 71.79%, respectively (area under ROC = 0.7662). Furthermore, the lymph node metastasis and TNM staging of patients were positively correlated with high expression of circGAPVD1. Combined with the ENCORI database and GEO datasets, we identified the circGAPVD1-related ceRNA network. The enrichment analysis revealed that key nodes in the ceRNA network participate in many important signaling pathways such as protein post-translational modifications
Conclusion
Our results revealed the diagnostic efficiency of circGAPVD1 in plasma exosomes. The highly expressed circGAPVD1 is expected to be a novel diagnostic marker for CRC.
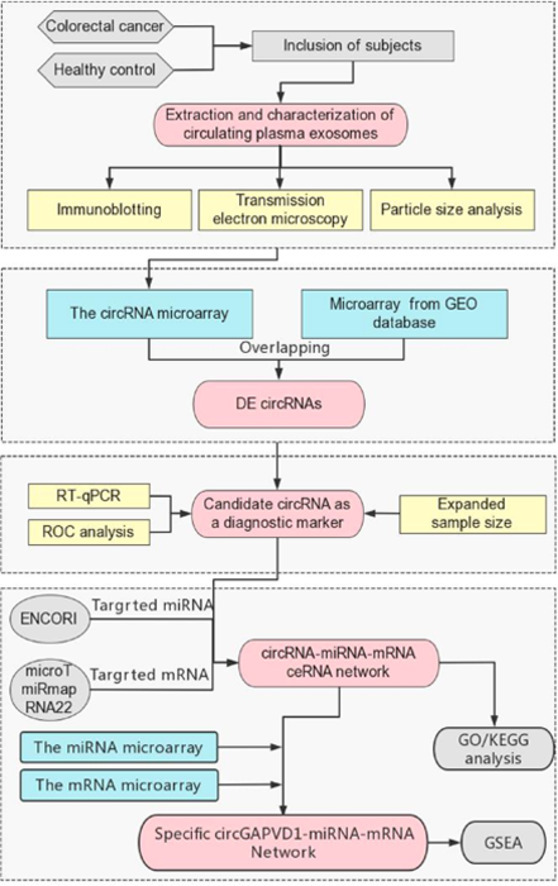
Graphical abstract
Work flowchart of the study.
Abbreviations
- CRC
colorectal cancer
- EVs
extracellular vesicles
- TEM
Transmission electron microscopy
- DLS
Dynamic light scattering
- qRT-PCR
Real-Time Quantitative Reverse Transcription PCR
- WB
Western Blot
- ROC
Receiver operating characteristic
- AUC
area under the ROC curve
- GO
Gene ontology
- KEGG
Kyoto Encyclopedia of Genes, Genomes
- GSEA
Gene set enrichment analysis
- CI
Confidence interval
- circRNAsCircular RNA miRNA
Circular RNA miRNAMicroRNA
- cDNA
Complementary DNA
- gDNA
genomic DNA
- MF
Molecular functions
- BP
Biological pathways
- CC
Cellular components
- CI
Confidence interval
- CLU
clusterin%2C transcript variant 1
- FOS2
FOS-like antigen 2
- GALNT6
polypeptide N-acetylgalactosaminyltransferase 6
- NOP2
NOP2 nucleolar protein%2C transcript variant 5
- PTMs
Protein post-translational modifications
Introduction
Colorectal cancer (CRC) is a common and high-malignancy tumor of the gastrointestinal system and shows the 2nd highest mortality and 3rd highest morbidity rates among common malignancies [1]. Early diagnosis and early treatment are vital for improving the overall survival rate of patients with CRC [2], [3], [4]. Although the currently used diagnostic approach of colonoscopy provides high accuracy, it has some risks in clinical application as it is an invasive test. Hence, there is a need to develop a safer and more effective tool for the clinical diagnosis and treatment of CRC [5,6].
Exosomes, which are a class of lipid bilayer-enclosed extracellular vesicles (EVs) of approximately 40–160 nm size, exist in a variety of biological fluids and can be secreted by almost all cells [5,7]. These small nanoscale vesicles differ from other EVs. Exosomes begin as intracellular outgrowths and then form multivesicular vesicles that fuse with the cell membrane [8]. Thereafter, they are exocytically released into the extracellular space. Exosomes are crucial mediators of intercellular transfer of information [7,9]. They are heterogenous in nature and include a variety of proteins, lipids, DNA, and RNA [10]. The transmembrane proteins CD63 and CD9; heat shock proteins HSP70 and HSP90; and tumor susceptibility gene 101 protein (TSG101) have been commonly used as biomarkers of exosomes [7,11,12]. Increasing evidence indicates that exosome-derived RNA molecules are involved in the development and progression of cancer and can be used as valid biomarkers for cancer [5,13].
CircRNA is a special class of RNA molecules that are single-stranded covalently closed loops lacking free 5′ and 3′ ends [14]. Compared to linear RNAs, circRNAs are difficult to degrade by external RNA enzymes due to their covalently closed ring structure, which allows them to exist stably in various cellular and external environments [15,16]. Exosomal circRNAs (exo‐circRNAs) extracted from plasma have excellent potential to be used as diagnostic biomarkers for various cancers as they show cell-specific enrichment and are relatively stable and evolutionarily conserved [17,18]. Compared with the specificity and sensitivity of CEA and CA19‐9 proteins to detect gastric cancer, the diagnostic specificity and sensitivity of hsa_circ_0001017 levels in the blood are higher [19]. Similarly, hsa_circ_0000826 shows a good diagnostic ability in CRC [20]. Pan et al. have shown that exo-circRNAs are upregulated in the serum of patients with CRC in comparison to that in patients with benign intestinal disease [21]. Thus, circRNAs and plasma exo-circRNAs are high‐efficient blood‐based biomarkers with great clinical significance [22]. However, at present, only a limited number of circRNAs have been studied, and even fewer exo‐circRNAs have been studied as biomarkers in CRC [23]. Therefore, the investigation of differentially-expressed circRNAs with important biological functions in CRC can provide new targets and insights for the diagnosis of CRC.
The current study aimed to detect novel early diagnostic biomarkers in the exosomes of patients with CRC. Briefly, we screened the upregulated critical exo-circRNAs extracted from the plasma of patients with CRC, constructed a ceRNA network, identified the differentially-expressed genes (DEGs) and their associated pathways, and validated the results by further analysis. The findings of this study are expected to provide a basis for not only the diagnosis of CRC but also for its prognosis and the development of molecular targeted therapy.
Materials and methods
Subjects
We conducted the retrospective case-control research by enrolling a total of 78 CRC patients and matched 78 healthy controls. All CRC patients were confirmed by postoperative histopathology in the affiliated Hospital of Xuzhou Medical University ranging from June 2020 to August 2021. All preoperative plasma samples were collected from untreated patients. Controls were obtained from healthy volunteers who performed health check-ups. All participants signed informed consent. This study was approved by an institutional review board of the affiliated Hospital of Xuzhou Medical University (XYFY2019-KL221–01) .
The inclusion criteria for the CRC group were determined with the following: 1. Patients with CRC who underwent surgery in our hospital 2. No pre-operative neoadjuvant radiotherapy or other special treatment 3. Complete pathological results after surgery; Exclusion criteria were determined with the following: 1. Pre-operative neoadjuvant radiotherapy 2. Unclear pathological diagnosis 3. Combined with other types of tumors; Inclusion criteria for the normal control group were determined with the following: people in good health and without special diseases.
Methods
Plasma collection
Before the procedure, peripheral venous blood (5 mL) was collected from each patient into an EDTA anticoagulation tube. After centrifugation at 2000 rpm for 10 min, samples were transferred to 1.5 mL RNase-free centrifuge tubes and stored at −80 °C.
Exosome extraction
Exosomes were isolated from plasma using the Invitrogen™ Total Plasma Exosome Isolation Kit (Cat: 4,484,450) according to the manufacturer's product instructions. For plasma samples, 250 μL of 1xPBS buffer was added to 500 μL of plasma sample, mixed it up, 25 μL of Proteinase K was added and then allowed to stand at room temperature for 10 min. 150 μL of Exosome Isolation Reagent was added, mixed it up, and then allowed to stand at 4 °C for 30 min. Then the supernatant was removed to obtain the exosome precipitate, and the exosome precipitate was resuspended in 150 μL of 1xPBS buffer.
Exosome identification
Transmission electron microscopy shoot: 20 μL of resuspended exosomes were taken onto the sealing film, the carbon film copper was placed on the exosome droplet, left for 10 min, filter paper was used to absorb water, the carbon film copper was transferred to 3% glutaraldehyde solution, left for 5 min and absorbed water to dry up, then transferred to a double-distilled water droplet and washed repeatedly 10 times for 2 min each, the washed carbon film copper was transferred to 4% hydrogen peroxide acetate solution. Stand for 30 s, aspirate the negative staining solution, dried at room temperature for 5 min. The images were obtained using transmission electron microscopy.
Western Blot analysis: Western Blot was used to detect the biomarker detection of exosomes, and the extracted exosomes were directly tested for protein concentration by BCA kit(Beyotime, Shanghai, China, Cat: E112–02). The obtained protein lysates were put into a 10% SDS-PAGE gel, transferred onto PVDF membranes (Millipore, Massachusetts, USA,Cat: IPVH00010). Following soaked in 5% skimmed milk, the PVDF membranes were incubated with primary antibodies specific against:CD63 (Proteintech, Chicago, IL, USA, Cat: 25,682–1-AP), TSG101 (Proteintech,Chicago,IL,USA,Cat:14,497–1-AP), Calnexin(Proteintech,Chicago,IL,USA, Cat: 10,427–2-AP) at 4 °C overnight. The day after, the PVDF membranes were incubated with the corresponding secondary antibody for 2 h at room temperature before washed with PBS-Tween 20 (PBST) buffer 3 times. Finally, Chemistar™ High-sig ECL Western Blot Substrate (Tanon, Shanghai, China,Cat: P10200) was employed to detect the blots.
Particle size analysis: Resuspended plasma exosomes were placed into a cuvette by dynamic light scattering (DLS), and their particle size distribution was measured and recorded using a Malvern dynamic light scattering instrument.
Screening of candidate circRNAs
After searching for CRC-related circRNA expression profiles in the GEO database, the GSE126094 dataset was finally selected as a further screening set, and the raw data were imported into R language software to screen the up-regulated expression of circRNAs with P<0.05, FC>2. In the meantime, three pairs of CRC patients and healthy subjects were commissioned Shanghai Kangchen Bio-tech Co. to perform deep sequencing of circRNA microarray data and perform analysis of circRNA microarray data with the same screening criteria as above. Finally, the intersection of circRNAs that were upregulated in both groups was selected for further validation.
Total RNA extraction from exosomes and real-time fluorescence quantitative PCR
Total RNA was isolated from the exosomes using RNA Isolater Total RNA Extraction Reagent (Vazyme, Nanjing, China,Cat:R401–01), and the concentration and purity of RNA were determined by UV spectrophotometer. The extracted RNA was considered pure when the A260/A280 ratio was between 1.8–2.0. qRT–PCR was then performed with miRNA Universal SYBR qPCR Master Mix(Vazyme, Nanjing, China,Cat:CW0957H) on a LightCycler 96 Instrument using the following cycling conditions: 95 °C for 5 min; 40 cycles at 95 °C for 10 s and 60 °C for 60 s; and a melting curve analysis. GAPDH serving as a reference for mRNAs.The relative RNA expression level was calculated using the 2-ΔΔCT method. The primers were designed and synthesized by Shanghai Biotech.
Ring formation validation of circGAPVD1
To verify the loop-forming property of circGAPVD1, we designed reverse and opposite specific primers that can specifically amplify the corresponding linear RNA and 18S corresponding sequences. The gDNA was extracted according to the steps of Tiangen Blood/Cell/Tissue Genomic DNA Extraction Kit(Cat: DP304), and then PCR amplification was performed using the extracted gDNA and reverse transcribed cDNA as templates. Finally, the PCR products were subjected to agarose gel electrophoresis, after electrophoresis, the agarose gel was put into the gel imaging analysis system. Finally, the size and position of the bands were photographed to observe.
Data acquisition and processing
CircRNA microarray dataset (GSE126094), miRNA microarray dataset (GSE126093), and mRNA microarray dataset (GSE126092) were downloaded from Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/). By using data tables of microarray platforms with R software, the probe IDs of the original data were converted to official gene symbols. Raw expression values were log 2 transformed with Aaffy package encoded by R. The normalize Between Arrays function of the R package limma used to normalize gene expression values.
Differential expression analysis of miRNAs, and mRNAs
The analysis of differentially expressed of miRNAs and mRNAs were performed using “limma” package in R software (4.1.2). The differentially expressed differentially expressed miRNAs (DEMs) and differentially expressed genes (DEGs) were screened with P-value 〈 0.05 and |logFC| 〉 1 as the threshold. Heatmaps and box line diagram of DE circRNAs and DE miRNAs were visualized using the “pheatmap” and “ggpubr” packages of R software, respectively.
Construction of the circRNA-miRNA-mRNA regulatory network
The target miRNAs of circRNAs were obtained using the Encyclopedia of RNA Interactomes (ENCORI, http://starbase.sysu.edu.cn/index.php). Furthermore, GEO databases were considered as candidate miRNAs and mRNAs to construct the ceRNA network. Then, the microT, miRmap, and RNA22 were used to predict the relationships among miRNAs and their target mRNAs. The visualization of regulatory network was performed by cytoscape 3.9.1.
Functional enrichment analysis of mRNAs
Functional Gene Ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were performed respectively. Enrichment analysis and drawing of graphs and of GO and KEGG pathways were performed using the “clusterProfiler”, “ggplot2”, and“org.Hs.eg.db” packages in R. GO terms and KEGG pathways were filtered at P < 0.05. Then, We divided CRC samples into two phenotype subgroups on the grounds of the median expression level of DE mRNAs. According to the degree of divergence between two subgroups, genes from the GEO expression profiles were ranked in a list, through GSEA software 4.1.
Statistical methods
Statistical analyses were performed using SPSS 20.0 software(IBM, Armonk, NY, USA) and the mean±standard deviation was used to express the normally distributed measures. Student's t-test was used to evaluate the significance of differences between groups. The chi-square test was used for clinicopathological characterization. The diagnostic performance of circRNA was evaluated by plotting ROC curves and calculating the AUC. A P value of < 0.05 was considered statistically significant.
Results
Extraction and characterization of circulating plasma exosomes in CRC and control group
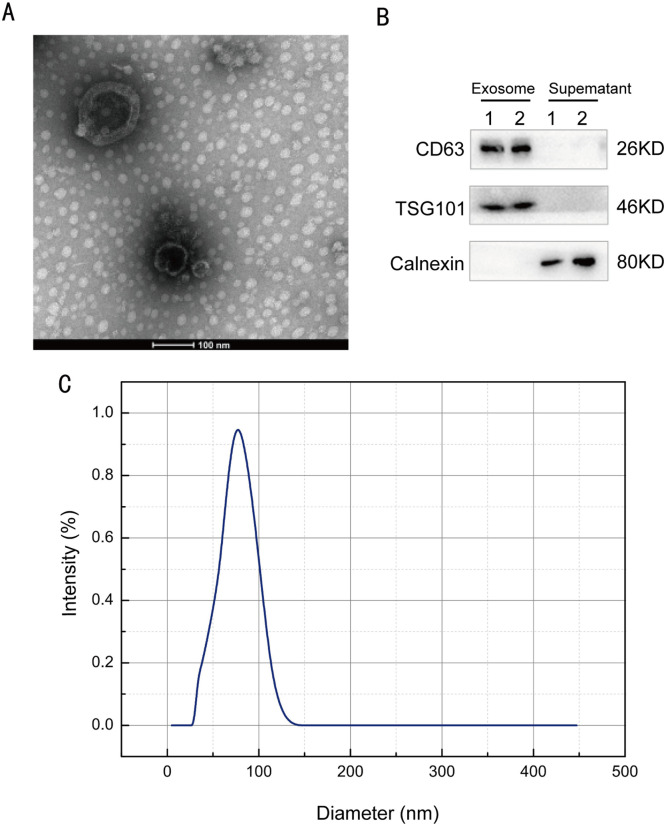
To explore the potential of plasma exo-circRNAs as diagnostic biomarkers for CRC, we first extracted exosomes from the plasma of patients with CRC and healthy control individuals using a Plasma Exosome Isolation Kit. Next, we examined the morphological appearance of the extracted exosomes by transmission electron microscopy (TEM). As shown in Fig. 1, the exosomes had a typical cup-holder structure (Fig. 1A). Western blotting (WB) showed the expression of the commonly used exosomal markers CD63 and TSG101 in the extracted exosomes, while the expression of the exosome negative marker calnexin was detected only in the supernatant fraction (Fig. 1B). Dynamic light scattering (DLS) showed that the diameter of the exosomes ranged between 40 and 160 nm (Fig. 1C). The morphological characteristics, biomarker expression, and size distribution of the extracted exosomes were all similar to those reported previously, indicating the successful extraction of exosomes.
Fig. 1.
Identification of circulating plasma exosomes in CRC and control groups.(A) A representation of transmission electron microscopy (TEM) images of plasma-derived exosomes. (B) Western blotting (WB) was used to detect the expression of CD9, TSG101, and calnexin in exosomes isolated from plasma samples of patients with CRC and healthy controls. (C) Dynamic light scattering (DLS) analysis results show the exosome diameters.
Screening of differentially-expressed circRNAs between CRC and control groups
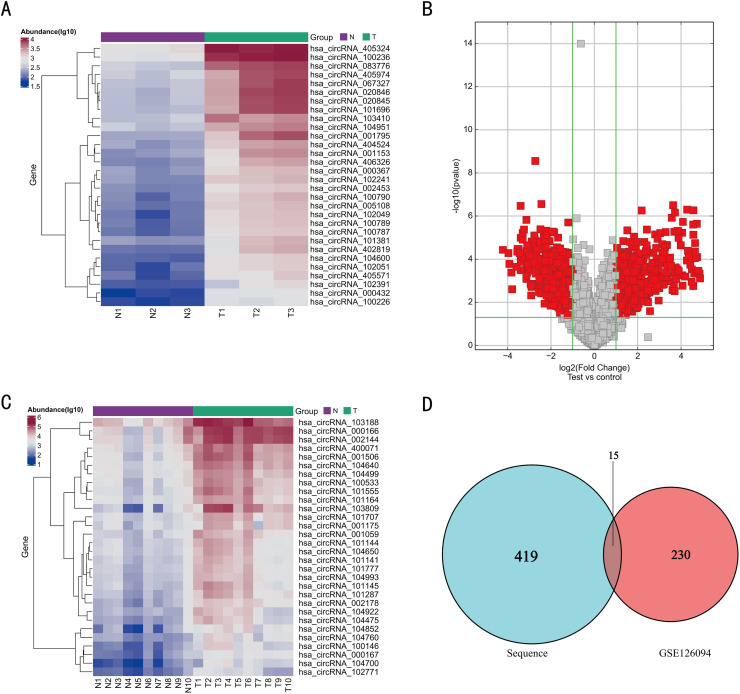
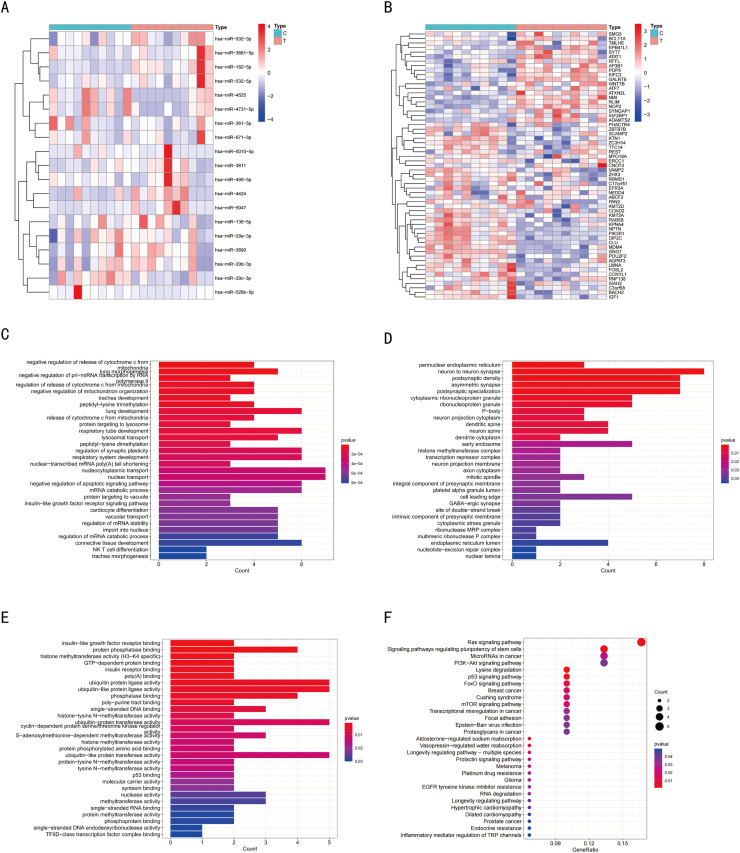
To screen for the differentially-expressed plasma exo-circRNAs in patients with CRC compared with that in healthy controls, we performed deep sequencing of three pairs of test samples and obtained a total of 434 differentially-expressed and up-regulated circRNAs with P-value < 0.05 and LogFC >2 as the cut-off set in the “limma” package in R software (version 4.1.2) (Fig. 2A, B). The heatmap shows the top 30 differentially-expressed and up-regulated circRNAs. To eliminate other potential factors influencing the circRNA levels in plasma-derived exosomes, we focused on the secretion of circRNAs from tumors into the blood. Therefore, we downloaded the CRC-related circRNA database (GSE126094) from the Gene Expression Omnibus (GEO) database, which comprised ten pairs of tumor tissues versus normal tissues, and subjected this microarray to differential analysis. As a result, 245 differentially-expressed and up-regulated circRNAs were screened with P-value < 0.05 and LogFC >2. The heatmap shows the top 30 circRNAs (Fig. 2C). Thereafter, the two microarray datasets were intersected, which revealed 15 circRNAs that exhibited differential expression (Fig. 2D). Supplementary Table 1 lists the specific circRNAs and their corresponding primer sequences.
Fig. 2.
CircRNA expression profile in CRC and control group and screening of differentially-expressed circRNAs.(A) Cluster analysis of the expression of the circRNAs by deep sequencing. The result shows the top 30 differentially-expressed circRNAs. (B) Volcano plot showing fold differences in gene expression. The red dots represent the upregulated or downregulated genes. (C) Cluster analysis of the differentially-expressed circRNAs in the GSE126094 dataset. The result shows the top 30 circRNAs. (D) Venn diagram of the differentially-expressed circRNAs obtained by clustering analysis. The data were analyzed with P-value < 0.05 and LogFC >2 as the threshold. Cluster analysis and Volcano plot analysis were performed using the gplots package in R.
Expression level of circGAPVD1 and its efficacy as a diagnostic marker
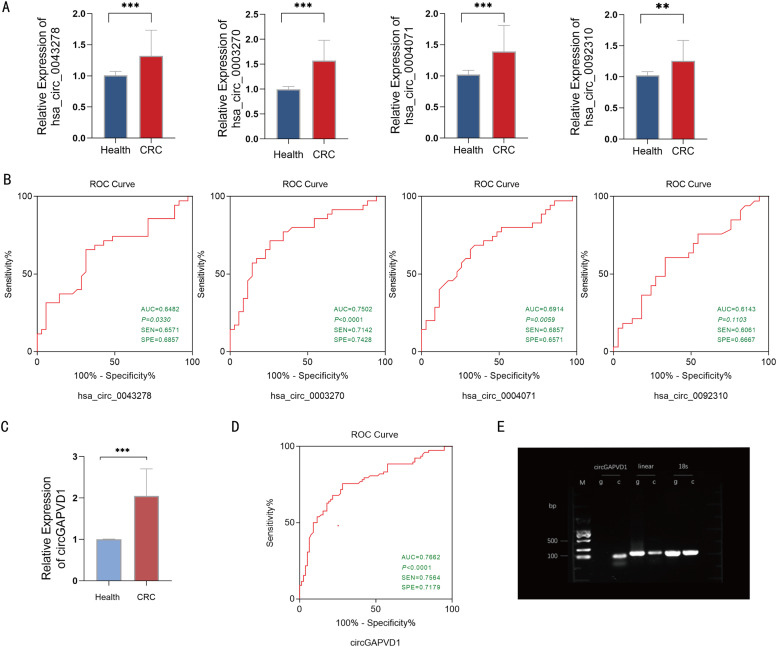
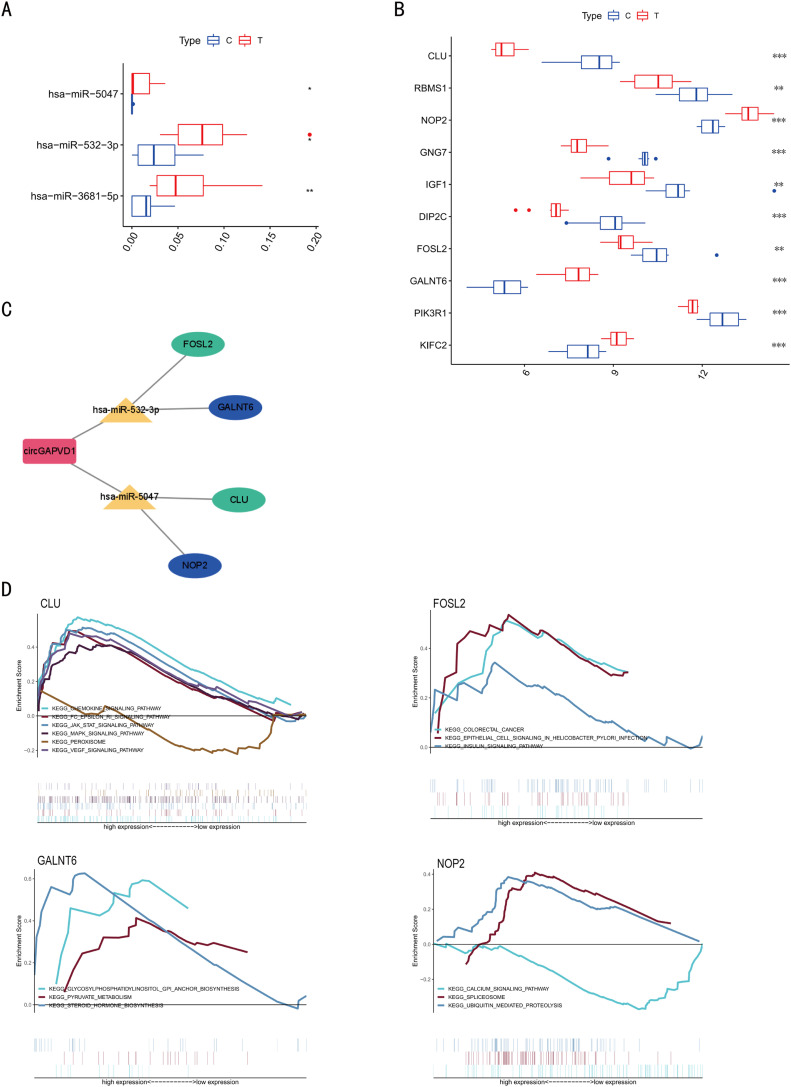
Multistage validation was performed for the 15 identified circRNAs. We randomly selected 35 patients with a pathological diagnosis of CRC and 35 healthy controls as the test group for the first batch of screening using plasma exosomes. Real-time quantitative PCR (qRT-PCR) was used to verify the differentially-expressed circRNAs in the CRC samples. The expression levels of four circRNAs, namely hsa_circ_0043278 (circTADA2A), hsa_circ_0003270 (circGAPVD1), hsa_circ_0004071 (circMYO9B), and hsa_circ_0092310 (circTJAP1), were significantly higher in the CRC group than in the control group (p<0.05 for all; Fig. 3A). Of these, circGAPVD1 showed the maximum variance and minimum p-value (P<0.001).
Fig. 3.
Expression level of circGAPVD1 and its efficacy as a diagnostic marker (A) Differential expression of four circRNAs in 35 paired CRC and healthy controls. The graphs were generated using GraphPad Prism software (version 8.0). (B) ROC curve analysis for the diagnostic ability of the four circRNAs in 35 paired CRC and healthy controls. (C) Differential expression of circGAPVD1 in 78 paired CRC and healthy controls with P-value < 0.001. (D) ROC curve analysis for the diagnostic ability of circGAPVD1 in 78 paired CRC and healthy controls. (E) The expression of circGAPVD1 examined using PCR and agarose gel electrophoresis. circGAPVD1 could be amplified by divergent primers in the complementary DNA (cDNA) but not genomic DNA (gDNA). 18S rRNA was used as a positive control. Results are shown as mean ± s.d, *p<0.05; **p < 0.01, ***p < 0.001, n.s., no significance.
To further analyze the diagnostic ability of the four circRNAs to distinguish patients with CRC from normal healthy individuals, the receiver operating characteristic (ROC) curve of the circRNAs for CRC was used to calculate the specific sensitivity and specificity. The area under the ROC curve (AUC) of hsa_circ_0043278, hsa_circ_0003270, hsa_circ_0004071, and hsa_circ_0092310 was 0.6482 (95% CI: 0.5175–0.7788, P = 0.0330), 0.7502 (95% CI: 0.6339–0.8665, P< 0.001), 0.6914 (95% CI: 0.5667–0.8162, P = 0.0059), and 0.6143 (95% CI: 0.4778–0.7508, P = 0.1103), respectively. CircGAPVD1 showed the highest performance, with a sensitivity of 71.42% and a specificity of 74.28% (Fig. 3B). Therefore, circGAPVD1 was selected for further studies.
Next, we expanded the number of clinical samples to a total of 78 pairs using the above criteria. The differential expression of circGAPVD1 in the 78 paired CRC and healthy control samples and the ROC of circGAPVD1 were analyzed. qRT-PCR analysis demonstrated a statistically significant difference in circGAPVD1 expression between the two groups (Fig. 3C). The AUC of circGAPVD1 was 0.7662 (95% CI:0.6908–0.8416, P< 0.0001), with a sensitivity of 75.64% and a specificity of 71.79% (Fig. 3D). This indicates that the level of plasma exosome-derived circGAPVD1 has a high diagnostic efficacy.
We then conducted the loop formation assay to verify the loop structure of circGAPVD1, and found that circGAPVD1 could be amplified by divergent primers using complementary DNA (cDNA) but not genomic DNA (gDNA), while its corresponding linear RNA and the 18S linear fragment could be obtained by the amplification of both cDNA and gDNA. This indirectly confirmed the loop structure of circGAPVD1 (Fig. 3E).
Relationship between circGAPVD1 levels and clinicopathological characteristics of patients with CRC
The median expression level of circGAPVD1 was used as the cut-off value to divide the CRC group into low-expression and high-expression groups. Such categorization revealed that high expression levels of circGAPVD1 were significantly correlated with the TNM stage and lymph node metastases in patients with CRC (P<0.05), while there was no significant association of circGAPVD1 levels with age, gender, tumor site, tumor size, or distant metastasis (Table 1).
Table 1.
Relationship between circGAPVD1 expression level and clinicopathological characteristics of CRC patients.
| Clinicopathological features |
Total number of cases (n = 78) |
circGAPVD1 expression level |
x2 | p-value | |
|---|---|---|---|---|---|
| Low expression (n = 39) | High expression (n = 39) | ||||
| Age (years) | |||||
| ≤65 | 52 | 28 | 24 | 0.923 | 0.337 |
| >65 | 26 | 11 | 15 | ||
| Gender | |||||
| Male | 45 | 23 | 22 | 0.053 | 0.819 |
| Female | 33 | 16 | 17 | ||
| Tumor site | |||||
| Colon | 34 | 15 | 19 | 0.834 | 0.361 |
| Rectum | 44 | 24 | 20 | ||
| Tumor size (cm) | |||||
| ≤5 | 53 | 28 | 25 | 0.530 | 0.467 |
| >5 | 25 | 11 | 14 | ||
| Lymph node metastasis | |||||
| N0 | 48 | 30 | 18 | 7.800 | 0.005 |
| N1–2 | 30 | 9 | 21 | ||
| Distant metastasis | |||||
| M0 | 74 | 38 | 36 | 1.054 | 0.305 |
| M1 | 4 | 1 | 3 | ||
| TNM Stage | |||||
| I–II | 47 | 29 | 18 | 6.478 | 0.011 |
| III–IV | 31 | 10 | 21 | ||
Construction of the ceRNA network and identification of the hub genes
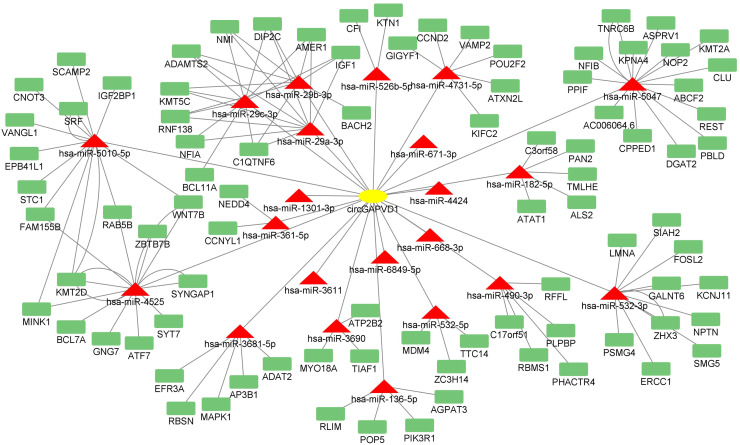
The role of circRNAs as miRNA sponges is the main mechanism of circRNA function in diseases. Therefore, we established a ceRNA network to explore the role of circGAPVD1. The circRNA-miRNA-mRNA ceRNA network was constructed utilizing circRNA-miRNA pairs and miRNA-mRNA pairs in combination. The ENCORI database prediction software was utilized to predict the downstream miRNAs of circGAPVD1 and the final regulated mRNAs. This database is superior to others in that the results can be filtered to identify target miRNAs that have been experimentally validated. The target genes of the target miRNAs were predicted simultaneously using microT, miRmap, and RNA22 in Starbase to construct a ceRNA network comprising 22 miRNAs and 88 mRNAs. The ceRNA network was plotted using Cytoscape 3.9.1 software (Fig. 4).
Fig. 4.
The specific circRNA-miRNA-mRNA ceRNA network.The competing endogenous RNA network was constructed based on the ceRNA theory. The yellow ellipse indicates circGAPVD1; the red triangle indicates miRNAs; the green triangle indicates mRNAs.
GO and KEGG function enrichment analysis
The gene expression profiles of miRNAs and mRNAs with the accession number GSE126093 and GSE126092 were downloaded from the GEO database, which contained 10 samples from patients with CRC and 10 healthy controls. The GSE126093 dataset was used for miRNA analysis and the GSE126092 was used for mRNA analysis. The results of mRNA and miRNA differential analysis are shown as heatmaps (Fig. 5A, B). Gene ontology (GO) and Kyoto Encyclopedia of Genes, Genomes (KEGG) enrichment analyses for mRNAs in the network were performed. The top 30 most significantly enriched biological processes (BPs), cell components (CCs), and molecular functions (MFs), with P-value < 0.05 as the cutoff criterion have been shown in Fig. 5C–E and Table 2. The top 30 terms were associated with multiple biological functions, including “regulation of release of cytochrome c from mitochondria”, “phosphatase binding”, and “ubiquitin-like protein ligase activity”. Additionally, KEGG pathway analysis (Fig. 5F, Table 3) revealed that the mRNAs were mainly enriched in pathways associated with “Ras signaling pathway” and “MicroRNAs in cancer”.
Fig. 5.
Functional enrichment analysis of mRNAs in ceRNA network. Cluster analysis of the differentially-expressed miRNAs (A) and mRNAs (B) in the GSE126093 and GSE126092 datasets with P-value < 0.05 and |LogFC| > 1. The top 30 most significantly enriched biological processes (C), cell components (D), and molecular functions (E) after GO analysis of differentially-expressed mRNAs in the ceRNA network with P-value < 0.05. (F) KEGG pathway enrichment analyses were performed using target genes in the ceRNA network. Enrichment was analyzed with P-value < 0.05. The color intensity of the nodes shows the degree of enrichment. The dot size represents the count of mRNAs in a pathway.
Table 2.
GO enrichment analysis for the differentially expressed genes in ceRNA network(top 30 and P-Value < 0.05).
| GO terms | GO entry | Count | P-value |
|---|---|---|---|
| nucleocytoplasmic transport | GO:0,006,913 | 6 | 0.000245625 |
| nuclear transport | GO:0,051,169 | 6 | 0.000245625 |
| establishment of protein localization to organelle | GO:0,072,594 | 6 | 0.001442753 |
| response to radiation | GO:0,009,314 | 6 | 0.002133975 |
| extrinsic apoptotic signaling pathway | GO:0,097,191 | 5 | 0.000452249 |
| negative regulation of apoptotic signaling pathway | GO:2,001,234 | 5 | 0.000501222 |
| mRNA catabolic process | GO:0,006,402 | 5 | 0.000587765 |
| negative regulation of cellular amide metabolic process | GO:0,034,249 | 5 | 0.001218901 |
| RNA catabolic process | GO:0,006,401 | 5 | 0.00132087 |
| regulation of mRNA metabolic process | GO:1,903,311 | 5 | 0.001543592 |
| regulation of protein stability | GO:0,031,647 | 5 | 0.00179288 |
| response to light stimulus | GO:0,009,416 | 5 | 0.002443975 |
| cellular response to abiotic stimulus | GO:0,071,214 | 5 | 0.002827355 |
| cellular response to environmental stimulus | GO:0,104,004 | 5 | 0.002827355 |
| negative regulation of organelle organization | GO:0,010,639 | 5 | 0.003503224 |
| regulation of apoptotic signaling pathway | GO:2,001,233 | 5 | 0.003858601 |
| peptidyl-lysine modification | GO:0,018,205 | 5 | 0.004859549 |
| nucleobase-containing compound catabolic process | GO:0,034,655 | 5 | 0.006757979 |
| synapse organization | GO:0,050,808 | 5 | 0.008150538 |
| heterocycle catabolic process | GO:0,046,700 | 5 | 0.009732034 |
| cellular nitrogen compound catabolic process | GO:0,044,270 | 5 | 0.010272612 |
| histone modification | GO:0,016,570 | 5 | 0.01141527 |
| aromatic compound catabolic process | GO:0,019,439 | 5 | 0.011814761 |
| positive regulation of catabolic process | GO:0,009,896 | 5 | 0.014530175 |
| organic cyclic compound catabolic process | GO:1,901,361 | 5 | 0.014882033 |
| ubiquitin protein ligase activity | GO:0,061,630 | 5 | 0.002302356 |
| ubiquitin-like protein ligase activity | GO:0,061,659 | 5 | 0.002721509 |
| ubiquitin-protein transferase activity | GO:0,004,842 | 5 | 0.010140186 |
| ubiquitin-like protein transferase activity | GO:0,019,787 | 5 | 0.012695759 |
| negative regulation of extrinsic apoptotic signaling pathway | GO:2,001,237 | 4 | 0.000188549 |
Table 3.
KEGG pathway analysis for the differentially expressed genes in ceRNA network (top 20 and P-Value < 0.05).
| KEGG pathway | KEGG entry | Count | P-value |
|---|---|---|---|
| Ras signaling pathway | hsa04014 | 5 | 0.00150489 |
| Signaling pathways regulating pluripotency of stem cells | hsa04550 | 4 | 0.001715907 |
| MicroRNAs in cancer | hsa05206 | 4 | 0.025390161 |
| PI3K-Akt signaling pathway | hsa04151 | 4 | 0.038726252 |
| Lysine degradation | hsa00310 | 3 | 0.001518792 |
| p53 signaling pathway | hsa04115 | 3 | 0.002320839 |
| FoxO signaling pathway | hsa04068 | 3 | 0.011847299 |
| Breast cancer | hsa05224 | 3 | 0.016137167 |
| Cushing syndrome | hsa04934 | 3 | 0.018570745 |
| mTOR signaling pathway | hsa04150 | 3 | 0.018888609 |
| Transcriptional misregulation in cancer | hsa05202 | 3 | 0.032799945 |
| Focal adhesion | hsa04510 | 3 | 0.036357529 |
| Epstein-Barr virus infection | hsa05169 | 3 | 0.036815855 |
| Proteoglycans in cancer | hsa05205 | 3 | 0.03820893 |
| Aldosterone-regulated sodium reabsorption | hsa04960 | 2 | 0.007978151 |
| Vasopressin-regulated water reabsorption | hsa04962 | 2 | 0.011154062 |
| Longevity regulating pathway - Multiple species | hsa04213 | 2 | 0.021407799 |
| Prolactin signaling pathway | hsa04917 | 2 | 0.026851559 |
| Melanoma | hsa05218 | 2 | 0.02829165 |
| Platinum drug resistance | hsa01524 | 2 | 0.029023222 |
Construction of the specific circGAPVD1-miRNA-mRNA network and gene set enrichment analysis
The relative gene expression levels of the key miRNAs and mRNAs in the ceRNA network were verified by comparing the data of the 10 paired CRC and healthy controls in the GSE126093 and GSE126092 datasets. The expression levels of the top 3 and top 10 differentially-expressed miRNAs and mRNAs in the ceRNA network, respectively, are shown in Fig. 6A, B. Interestingly, two miRNAs (hsa-miR-532–3p, hsa-miR-5047) and four mRNAs (GLU, FOSL2, GALNT6, NOP2) formed a specific ceRNA network with circGAPVD1 (Fig. 6C). The four mRNAs were analyzed through gene set enrichment analysis (GSEA) to identify signaling pathways that were differentially activated in CRC between the low- and high-expression groups (Fig. 6D). According to the normalized enrichment score (NES) and P value, GSEA results suggested that GLU was most significantly enriched in “chemokine signaling pathway”, and “JAK-STAT signaling”. Meanwhile, FOSL2 might participate in “colorectal cancer”, GALNT6 might associate with “steroid hormone biosynthesis”, and NOP2 might be involved in “spliceosome” and “ubiquitin-mediated proteolysis”.
Fig. 6.
Construction of circGAPVD1-miRNA-mRNA regulatory network and Gene Set Enrichment Analysis.(A) Differential expression of three miRNAs in 10 paired CRC and healthy controls in the GSE126093 dataset. (B) Differential expression of 10 mRNAs in 10 paired CRC and healthy controls in the GSE126092 dataset. Data are presented as a plot of the median and range of relative expression levels. *p<0.05; **p < 0.01, ***p < 0.001. (C) circGAPVD1-miRNA-mRNA regulatory network. (D) GSEA of key genes based on KEGG gene sets.
Discussion
The sensitivity and specificity of the commonly used CRC markers CEA and CA19–9 proteins are inadequate, as some patients with CRC are diagnosed at an already advanced stage [24,25]. Although colonoscopy, an invasive endoscopic examination method, can directly view the lesion under the microscope, making biopsy and treatment possible, there are certain associated limitations and postoperative complications [26]. Accurate liquid biopsy is desirable nowadays as it can be utilized for the diagnosis and prevention of diseases through the analysis of specific components of the organism [27]. It has great prospects in clinical application owing to its many advantages such as less trauma, easy operation, the possibility of repeated sampling, and low cost [28].
Exosomes with a lipid bilayer membrane structure not only stabilize themselves in the circulatory system but also prevent the degradation of circRNAs by the external environment [23]. Exosomes derived from tumor cells can release circRNAs into the cellular or circulatory system in various ways [29]. Some circRNAs, such as circN4BP2L2 and circGSE1, are mainly present in plasma exosomes and are enriched and stable in exosomes [30]. Furthermore, the combination of the bilayer membrane structure of the exosomes and the covalently closed ring structure of circRNAs has led to a great interest in the study of exo-circRNAs in peripheral blood as disease biomarkers. For example, the expression of exo-circRNA IARS was reported to be higher in pancreatic cancer tissues and plasma exosomes compared to that in controls [31]. Moreover, the results of the study implied that the presence of exo-circRNA might function as a biomarker for the diagnosis and prognosis of pancreatic ductal adenocarcinoma. Another study demonstrated that circRNA_0001178 and circRNA_0000826 were significantly upregulated in CRC-m tissues, and both had the potential for differentially diagnosing liver metastases from CRC [20]. A growing number of studies have shown that exo-circRNAs have efficient diagnostic efficacy and broad research potential [32,33]. We anticipate that in the next five years, new targets and concepts will emerge that will help with the clinical translation and application of exo-circRNAs. Currently, however, there is no consistency in terms of detection, diagnosis, or therapy based on exo-circRNAs, despite the promise of liquid biopsy techniques utilizing non-coding RNA. Therefore, the development of standardized treatment protocols is eagerly waited.
In this study, to clarify whether circRNAs are truly altered in their expression due to tumor effects and to exclude alterations in functional circRNA expression, we intentionally screened tumor tissues and corresponding normal tissues from public databases. By analyzing and overlapping the results of two microarrays, we screened out fifteen circRNA molecules that could have diagnostic potential for CRC. Further, experimental verification by qRT-PCR revealed four circRNAs – hsa_circ_0043278, hsa_circ_0003270, hsa_circ_0004071, and hsa_circ_0092310 – that showed significantly different levels of expression between the CRC and control groups. Combining the qRT-PCR and ROC analysis results, circGAPVD1 was selected for further investigation.
Upon expanding the sample size to 78 paired volunteers, the qRT-PCR and ROC analyses showed similar results as before in terms of up-regulated expression and high diagnostic power of circGAPVD1. The AUC reached 0.7662 with a sensitivity of 75.64% and a specificity of 71.79%, which is superior to the previously reported AUCs for the commonly used markers CEA (0.682), CA19–9 (0.560), CA125 (0.590), and CA242 (0.651) [34]. Furthermore, correlation analysis showed that a high expression of circGAPVD1 in plasma exosomes was significantly correlated with the clinicopathological features of lymph node metastasis and TNM stage in patients with CRC (P < 0.05). Together, these findings suggest that the plasma exo-circRNA circGAPVD1 has high sensitivity and specificity as a diagnostic marker for CRC, and the high expression of circGAPVD1 is associated with some clinicopathological characteristics.
Since circRNAs contain a large amount of miRNA binding sites, they can act as miRNA molecular sponges and indirectly regulate the expression of downstream target genes of the miRNAs [35], [36], [37]. Rengganaten et al. [38] showed that ceRNAs (hsa_circ_0066631 and hsa_circ_0082096) may be related to the establishment and maintenance of stemness in reprogrammed CRC cancer stem cells, suggesting that circRNAs are ceRNAs with important biological significance.
Hence, we constructed a circRNA-miRNA-mRNA regulatory network using a bioinformatics analysis approach to elucidate the key role(s) of circGAPVD1 in the pathogenesis of CRC and thereby guide the diagnosis and treatment of CRC [39]. The CRC-related datasets GSE126093 and GSE126092 were downloaded from the GEO database and used for GO enrichment analysis, which indicated that the enriched mRNAs are involved in many biological processes such as “regulation of release of cytochrome c from mitochondria” and “phosphatase binding”. Additionally, the most common molecular functions included “ubiquitin-like protein ligase activity” and “phosphatase binding”. Most of the biological processes and major molecular functions are associated with protein post-translational modifications (PTMs) [40]. Previous studies have reported the association of PTMs with CRC and indicated their potential use in the early diagnosis of CRC [41,42]. The enrichment analysis was enriched in “ubiquitin-protein transferase activity”, “ubiquitin protein ligase activity”, and “protein phosphatase binding”. Previous studies also support the role of these pathways in CRC. For example, PTPRF acts as an oncogenic protein phosphatase that activates Wnt signaling in CRC [43]. Ubiquitin ligase is not only involved in protein localization, metabolism, regulation, and degradation but also in the regulation of cell cycle, proliferation, apoptosis, metastasis, gene expression, and transcriptional regulation [44,45]. Several studies have confirmed that this pathway is closely linked to the development of CRC [46,47].
To further understand the potential mechanisms of the four key mRNAs in the ceRNA network, we performed GSEA [48], which revealed that these differentially-expressed mRNAs are involved in several important signaling pathways. The prognosis and progression of CRC have been associated with “JAK-STAT signaling” [49], “steroid hormone biosynthesis” [50], and “ubiquitin-mediated proteolysis” [51]. Circ-LNLM1 blocks AKT ubiquitination in patients with lymph node-negative CRC with synchronous liver metastases, indicating that it may be a prognostic and diagnostic biomarker for CRC [52]. In conclusion, the key mRNAs in the ceRNA network could be involved in CRC development by regulating relevant signaling pathways.
Although our results provide a basis for further mechanistic research, our study has several limitations. First, the sample size was relatively small. Therefore, the verification of exosomal circGAPVD1 as a diagnostic and prognostic marker using larger CRC cohorts is necessary for its application in early clinical diagnosis of patients. Second, we did not investigate the diagnostic potential of the remaining candidate circRNAs. Nonetheless, our study demonstrates the clinical importance of circGAPVD1 as a diagnostic marker for the first time. We also predict its targeted miRNAs and mRNAs to explore plausible and important functions, thereby opening new research avenues that can have ramifications for the early clinical diagnosis of CRC and clinical treatment.
Conclusions
In summary, our study compared the differences in the expression levels of circRNAs in plasma exosomes between CRC and healthy populations and evaluated the clinical application value of circGAPVD1. Our analyses suggest that plasma exosomal circGAPVD1 can be a potential novel biomarker for the diagnosis and dynamic monitoring of CRC through a minimally-invasive method. Furthermore, we also identified its targeted miRNAs and mRNAs, which may play a significant and potential role in CRC.
Authors'contributions
All authors agree to publication of the article.
Ethics approval and consent to participate
This study was approved by the Ethical Review at the Affiliated Hospital of Xuzhou Medical University.
Funding
This research was supported by grants from the National Natural Science Foundation of China (No. 82072729), the Natural Science Foundation of Jiangsu (BK20211606), Xuzhou Key R&D Program (KC20064) and Youth Science and Technology Fund of China University of Mining and Technology (2020QN79).
Declaration of Competing Interest
There are no competing interests between all authors.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2023.101652.
Contributor Information
Xin Liu, Email: 489507198@qq.com.
Yi Zhang, Email: dr_zhy@xzhmu.edu.cn.
Appendix. Supplementary materials
References
- 1.Bray F., et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Shaukat A., et al. ACG clinical guidelines: colorectal cancer screening 2021. Am. J. Gastroenterol. 2021;116:458–479. doi: 10.14309/ajg.0000000000001122. [DOI] [PubMed] [Google Scholar]
- 3.Rizzo A., et al. Dose reduction and discontinuation of standard-dose regorafenib associated with adverse drug events in cancer patients: a systematic review and meta-analysis. Ther. Adv. Med. Oncol. 2020;12 doi: 10.1177/1758835920936932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.André T., et al. Pembrolizumab in microsatellite-instability-high advanced colorectal cancer. N. Engl. J. Med. 2020;383:2207–2218. doi: 10.1056/NEJMoa2017699. [DOI] [PubMed] [Google Scholar]
- 5.Yu D., et al. Exosomes as a new frontier of cancer liquid biopsy. Mol. Cancer. 2022;21:56. doi: 10.1186/s12943-022-01509-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baassiri A., et al. Exosomal non coding RNA in LIQUID biopsies as a promising biomarker for colorectal cancer. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21041398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalluri R., LeBleu V.S. The biology function and biomedical applications of exosomes. Science. 2020;367 doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeppesen D.K., et al. Reassessment of exosome composition. Cell. 2019;177 doi: 10.1016/j.cell.2019.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hingorani S.R. Intercepting cancer communiques: exosomes as heralds of malignancy. Cancer Cell. 2015;28:151–153. doi: 10.1016/j.ccell.2015.07.015. [DOI] [PubMed] [Google Scholar]
- 10.Isaac R., Reis F.C.G., Ying W., Olefsky J.M. Exosomes as mediators of intercellular crosstalk in metabolism. Cell Metab. 2021;33:1744–1762. doi: 10.1016/j.cmet.2021.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vlassov A.V., Magdaleno S., Setterquist R., Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Biophys. Acta. 2012;1820:940–948. doi: 10.1016/j.bbagen.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 12.Zaborowski M.P., Balaj L., Breakefield X.O., Lai C.P. Extracellular vesicles: composition, biological relevance, and methods of study. Bioscience. 2015;65:783–797. doi: 10.1093/biosci/biv084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu L., et al. Isolation and characterization of exosomes for cancer research. J. Hematol. Oncol. 2020;13:152. doi: 10.1186/s13045-020-00987-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z., et al. Tumor-suppressive circular RNAs: mechanisms underlying their suppression of tumor occurrence and use as therapeutic targets. Cancer Sci. 2019;110:3630–3638. doi: 10.1111/cas.14211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raza A., et al. Dynamic liquid biopsy components as predictive and prognostic biomarkers in colorectal cancer. J. Exp. Clin. Cancer Res. 2022;41:99. doi: 10.1186/s13046-022-02318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Memczak S., et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 17.Rybak-Wolf A., et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 18.Kristensen L.S., Jakobsen T., Hager H., Kjems J. The emerging roles of circRNAs in cancer and oncology. Nat. Rev. Clin. Oncol. 2022;19:188–206. doi: 10.1038/s41571-021-00585-y. [DOI] [PubMed] [Google Scholar]
- 19.Li T., et al. Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT-PCR detection. J. Mol. Med. 2018;96:85–96. doi: 10.1007/s00109-017-1600-y. [DOI] [PubMed] [Google Scholar]
- 20.Xu H., Wang C., Song H., Xu Y., Ji G. RNA-Seq profiling of circular RNAs in human colorectal Cancer liver metastasis and the potential biomarkers. Mol. Cancer. 2019;18:8. doi: 10.1186/s12943-018-0932-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan B., et al. Identification of serum exosomal hsa-circ-0004771 as a novel diagnostic biomarker of colorectal cancer. Front. Genet. 2019;10:1096. doi: 10.3389/fgene.2019.01096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y., Li Z., Xu S., Guo J. Novel potential tumor biomarkers: circular RNAs and exosomal circular RNAs in gastrointestinal malignancies. J. Clin. Lab. Anal. 2020;34:e23359. doi: 10.1002/jcla.23359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y., et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol. Cancer. 2019;18:116. doi: 10.1186/s12943-019-1041-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beermann J., Piccoli M.-T., Viereck J., Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol. Rev. 2016;96:1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 25.Dong D, et al. Identification of serum periostin as a potential diagnostic and prognostic marker for colorectal cancer. Clin. Lab. 2018;64:973–981. doi: 10.7754/Clin.Lab.2018.171225. [DOI] [PubMed] [Google Scholar]
- 26.Ogata-Kawata H., et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One. 2014;9:e92921. doi: 10.1371/journal.pone.0092921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cha J.M., et al. Risks and benefits of colonoscopy in patients 90 years or older, compared with younger patients. Clin. Gastroenterol. Hepatol. 2016;14 doi: 10.1016/j.cgh.2015.06.036. [DOI] [PubMed] [Google Scholar]
- 28.Marcuello M., et al. Circulating biomarkers for early detection and clinical management of colorectal cancer. Mol. Aspects Med. 2019;69:107–122. doi: 10.1016/j.mam.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 29.Bach D.-H., Lee S.K., Sood A.K. Circular RNAs in Cancer. Mol. Ther. Nucl. Acids. 2019;16:118–129. doi: 10.1016/j.omtn.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y., et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J., et al. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J. Exp. Clin. Cancer Res. 2018;37:177. doi: 10.1186/s13046-018-0822-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang M., He Y.-R., Liang L.-C., Huang Q., Zhu Z.-Q. Circular RNA hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer. World J. Gastroenterol. 2017;23:6330–6338. doi: 10.3748/wjg.v23.i34.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang M., Yu F., Li P., Wang K. Emerging Function and Clinical Significance of Exosomal circRNAs in Cancer. Mol. Ther. Nucl. Acids. 2020;21:367–383. doi: 10.1016/j.omtn.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo H., et al. Clinical significance and diagnostic value of serum NSE, CEA, CA19-9, CA125 and CA242 levels in colorectal cancer. Oncol. Lett. 2020;20:742–750. doi: 10.3892/ol.2020.11633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang Y., et al. The Roles of circRNAs in Liver Cancer immunity. Front. Oncol. 2020;10 doi: 10.3389/fonc.2020.598464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng Q., et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hansen T.B., et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 38.Rengganaten V., et al. Mapping a circular RNA-microRNA-mRNA-signaling regulatory axis that modulates stemness properties of cancer stem cell populations in colorectal cancer spheroid cells. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21217864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen L., Liu S., Tao Y. Regulating tumor suppressor genes: post-translational modifications. Signal Transduct. Target Ther. 2020;5:90. doi: 10.1038/s41392-020-0196-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu G., Jin L., Sun W., Wang S., Liu N. Proteomics of post-translational modifications in colorectal cancer: discovery of new biomarkers. Biochim Biophys. Acta Rev. Cancer. 2022;1877 doi: 10.1016/j.bbcan.2022.188735. [DOI] [PubMed] [Google Scholar]
- 42.Kopylov A.T., et al. Revelation of proteomic indicators for colorectal cancer in initial stages of development. Molecules. 2020;25 doi: 10.3390/molecules25030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gan T., et al. Inhibition of protein tyrosine phosphatase receptor type F suppresses Wnt signaling in colorectal cancer. Oncogene. 2020;39:6789–6801. doi: 10.1038/s41388-020-01472-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buetow L., Huang D.T. Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2016;17:626–642. doi: 10.1038/nrm.2016.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun T., Liu Z., Yang Q. The role of ubiquitination and deubiquitination in cancer metabolism. Mol. Cancer. 2020;19:146. doi: 10.1186/s12943-020-01262-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kathania M., et al. Itch inhibits IL-17-mediated colon inflammation and tumorigenesis by ROR-γt ubiquitination. Nat. Immunol. 2016;17 doi: 10.1038/ni.3488. [DOI] [PubMed] [Google Scholar]
- 47.Deng L., et al. Ubiquitination of Rheb governs growth factor-induced mTORC1 activation. Cell Res. 2019;29:136–150. doi: 10.1038/s41422-018-0120-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li X., et al. Validating expression and its prognostic significance in lung adenocarcinoma based on data mining and bioinformatics methods. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.720302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gordziel C., Bratsch J., Moriggl R., Knösel T., Friedrich K. Both STAT1 and STAT3 are favourable prognostic determinants in colorectal carcinoma. Br. J. Cancer. 2013;109:138–146. doi: 10.1038/bjc.2013.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li S., et al. Sex hormones and genetic variants in hormone metabolic pathways associated with the risk of colorectal cancer. Environ. Int. 2020;137 doi: 10.1016/j.envint.2020.105543. [DOI] [PubMed] [Google Scholar]
- 51.Yu L., et al. Ubiquitination-mediated degradation of SIRT1 by SMURF2 suppresses CRC cell proliferation and tumorigenesis. Oncogene. 2020;39:4450–4464. doi: 10.1038/s41388-020-1298-0. [DOI] [PubMed] [Google Scholar]
- 52.Tang J., et al. CircRNA circ_0124554 blocked the ubiquitination of AKT promoting the skip lymphovascular invasion on hepatic metastasis in colorectal cancer. Cell Death. Dis. 2021;12:270. doi: 10.1038/s41419-021-03565-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.