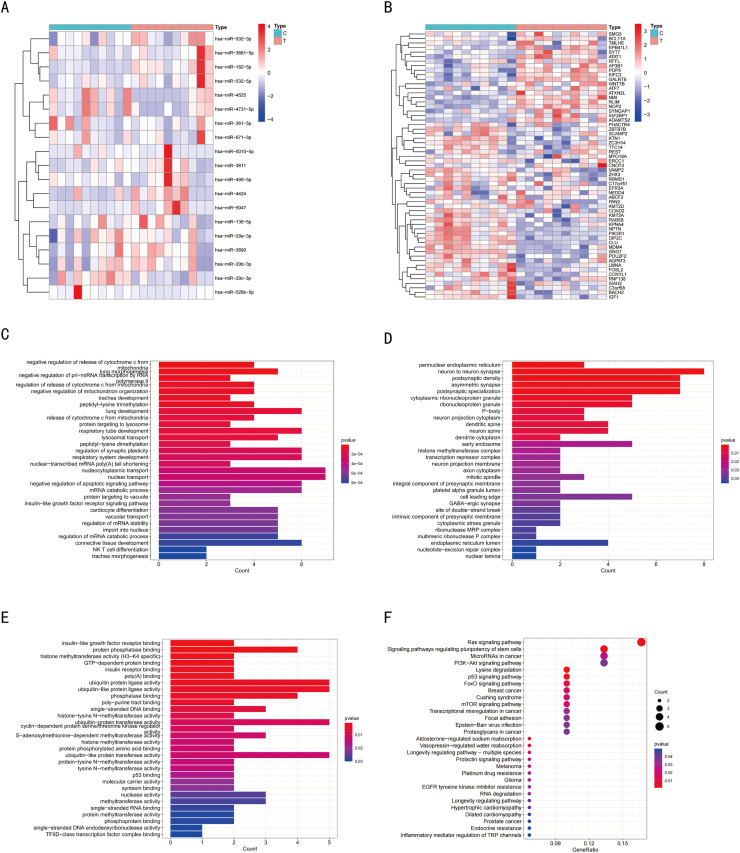
Fig. 5.
Functional enrichment analysis of mRNAs in ceRNA network. Cluster analysis of the differentially-expressed miRNAs (A) and mRNAs (B) in the GSE126093 and GSE126092 datasets with P-value < 0.05 and |LogFC| > 1. The top 30 most significantly enriched biological processes (C), cell components (D), and molecular functions (E) after GO analysis of differentially-expressed mRNAs in the ceRNA network with P-value < 0.05. (F) KEGG pathway enrichment analyses were performed using target genes in the ceRNA network. Enrichment was analyzed with P-value < 0.05. The color intensity of the nodes shows the degree of enrichment. The dot size represents the count of mRNAs in a pathway.