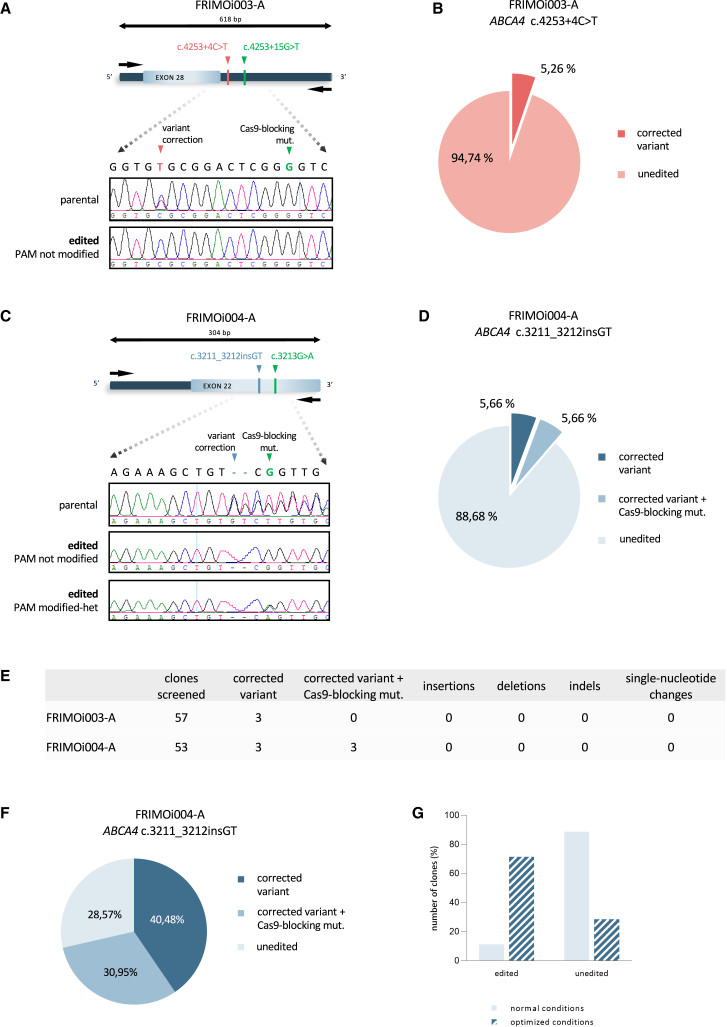
Figure 3.
Efficient correction of STGD1-related variants by CRISPR-Cas9 in hiPSCs
(A) FRIMO003-A hiPSC clones were subjected to Sanger sequencing to analyze gene editing in the locus specified after CRISPR-Cas9. Representative captures of chromatograms from Sanger results of parental and one edited clone are shown. (B) Pie chart of the percentage of edited clones in FRIMOi003-A cells. (C) As in (A) but with FRIMOi004-A. Representative capture of edited clones with PAM modification is also shown. (D) Pie chart of the percentage of edited clones according to variant correction and PAM modification in FRIMOi004-A cells. (E) Summary of the on-target events in total screened clones for each hiPSC line after CRISPR-Cas9-mediated gene editing. (F) Pie chart of the percentage of edited clones in FRIMOi004-A according to variant correction and PAM modification after cell culture conditions optimization. (G) Comparison between CRISPR-Cas9-mediated gene editing efficiency assays after normal and optimized conditions in the FRIMOi004-A hiPSC line.