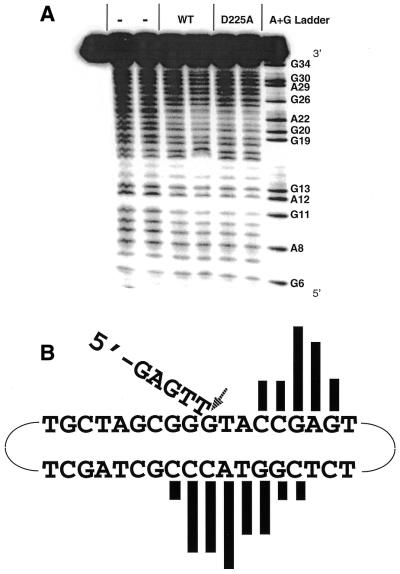
Figure 6.
The DNA footprint of Exo1 bound to the continuous 5′-flap substrate. (A) Hydroxyl radical footprinting reactions with Hex1-N2 and D225A mutant protein bound to 5′-flap DNA. A representative phosphorimager scan of three independent experiments is shown. The first lane (far left) is the no cleavage control. The – lanes are the no protein controls. Wild-type Hex1-N2 was used at 0.956 and 1.912 µg and the D225A Hex1-N2 mutant at 0.21 and 0.63 µg (from left to right). The A+G chemical sequencing ladder is shown, as are relevant nucleotide positions in the continuous 5′-flap substrate of (B). (B) Summary of the hydroxyl radical footprinting data. The filled vertical bars above or below each base indicate sites of protection from cleavage. The height indicates the relative strength of footprinting protection. The arrow indicates the site of endonuclease cleavage, between positions T5 and G6. While the data in this panel are the footprint of wild-type Hex1-N2, an essentially identical footprint was observed with the D225A Hex1-N2 mutant [see (A)].