Abstract
Identification of Nocardia and Mycobacterium species by matrix‐assisted laser desorption/ionization‐time of flight mass spectrometry (MALDI‐TOF MS) is still a challenging task that requires both suitable protein extraction procedures and extensive databases. This study aimed to evaluate the VITEK MS Plus system coupled with updated RUO (v4.17) and IVD (v3.2) databases for the identification of Nocardia spp. and Mycobacterium spp. clinical isolates. Sample preparation was carried out using the VITEK MS Mycobacterium/Nocardia kit for protein extraction. From 90 Nocardia spp. isolates analysed, 86 (95.6%) were correctly identified at species or complex level using IVD and 78 (86.7%) using RUO. Only two strains were misidentified as other species pertaining to the same complex. Among the 106 non‐tuberculous Mycobacterium clinical isolates tested from a liquid culture medium, VITEK MS identified correctly at species or complex level 96 (90.6%) isolates in the IVD mode and 89 (84.0%) isolates in the RUO mode. No misidentifications were detected. Although the IVD mode was unable to differentiate members of the M. fortuitum complex, the RUO mode correctly discriminated M. peregrinum and M. septicum. The robustness and accuracy showed by this system allow its implementation for routine identification of these microorganisms in clinical laboratories.
A standardized, MALDI‐TOF based, commercial method for the rapid identification of Nocardia and non‐tuberculous mycobacterium (NTM) isolates has been evaluated. The method allowed 95.6% and 90.6% correct species‐level identification for Nocardia and NTM, respectively.
INTRODUCTION
Nocardia spp. and Mycobacterium spp. are the causative agents of opportunistic infections in immunocompromised and immunocompetent individuals, mainly of pulmonary presentation but also cutaneous and soft‐tissue infections (Conville et al., 2018; Falkinham, 2016). Moreover, isolation of non‐tuberculous mycobacteria (NTM) has been increasing in last years in clinical microbiology laboratories (Alcaide et al., 2017). Rapid identification of clinical isolates of these two groups of microorganisms at species level is essential due to different antimicrobial susceptibility patterns among them (Conville et al., 2018; Griffith et al., 2007).
Identification of these two groups of microorganisms has been traditionally performed by phenotypic and biochemical methods, which have been replaced by molecular techniques (Sinner et al., 2015). In the case of non‐tuberculous Mycobacterium spp., they can be identified by commercial kits based on PCR‐reverse hybridization, but only for a limited number of NTM species (Makinen et al., 2006). DNA sequencing services are then required, which may reveal laborious and costly.
The use of matrix‐assisted laser desorption/ionization‐time of flight mass spectrometry (MALDI‐TOF MS) has been widely implemented for microbial identification (Patel, 2015). However, some acid‐fast microorganisms like Nocardia spp. and Mycobacterium spp. still represent a challenge. Due to the characteristics of their cell wall, they require a specific protein extraction procedure in order to obtain protein spectra suitable for identification by MALDI‐TOF MS. Mycobacteria species also require an inactivation protocol due to their pathogenicity. The use of a commercial extraction kit can allow both extraction an inactivation. Several studies have evaluated different protein extraction protocols for Nocardia spp. and Mycobacterium spp. isolates (Girard et al., 2017; Rodriguez‐Temporal et al., 2018; Saleeb et al., 2011). In addition, the updating of databases and implementation of in‐house libraries can improve identification rate of these microorganisms but in the case of in‐house databases, the results are not easy transposable to other laboratories (Rodriguez‐Sanchez et al., 2016; Rodriguez‐Temporal et al., 2017) (Marin et al., 2018).
The aim of this study was to evaluate the VITEK MS system using commercial and widely used In Vitro Diagnostic (IVD) and Research Use Only (RUO) databases for the identification of Nocardia spp. and Mycobacterium spp. with the help of a commercial kit for the protein extraction protocol.
EXPERIMENTAL PROCEDURES
Bacterial isolates
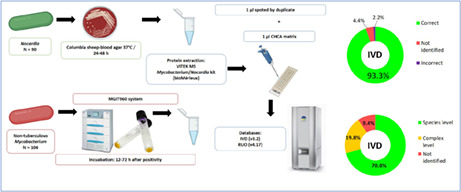
A total of 90 Nocardia isolates from 85 patients, representing 14 species (Table S1), and 106 non‐tuberculous Mycobacterium isolates from 100 patients, encompassing 21 species (Table S2), were included in the study. They all sourced from clinical samples that were submitted to the Department of Clinical Microbiology and Infectious Diseases of the Hospital General Universitario Gregorio Marañón (Madrid, Spain) for identification between 1995 and 2021, and frozen at −80°C. Nocardia spp. isolates were previously identified by 16S rRNA and/or secA1 gene sequencing, thawed and cultured on Columbia sheep‐blood agar (bioMérieux, Marcy l'Etoile, France) for 48 h at 37°C, the time required for colonies to be visible (Durand et al., 2020; Marin et al., 2018). Mycobacterium isolates were previously identified by 16S rRNA and/or hsp65 gene sequencing (Rodriguez‐Temporal et al., 2022), thawed and cultured in the BACTEC MGIT960 system following the manufacturer instructions (Becton Dickinson) until the tubes flagged positive.
Protein extraction and MALDI‐TOF MS analysis
All isolates were processed with the VITEK MS Mycobacterium/Nocardia kit (bioMérieux) following the manufacturer's instructions. For Nocardia spp. strains, colonies were harvested with a cytology brush and transferred to a 1.5 ml Eppendorf tube with 500 μl of 70% ethanol and 0.5 mm glass beads. Then, the tubes were vortexed for 15 min for inactivation, followed by 10 min incubation at room temperature and the suspension was transferred to an empty tube, discarding the glass beads. Samples were centrifuged at 18,800 g for 2 min and the supernatant was discarded. Ten microliters of 70% formic acid were added to the pellet and the tubes were vortexed for 10 s. Then, 10 μl of acetonitrile were added and the tubes were vortexed again. Finally, the tubes were centrifuged at 14,000 rpm for 2 min, and 1 μl of supernatant was spotted in duplicate on the MALDI‐TOF MS slide. After drying at room temperature, 1 μl of α‐cyano‐4‐hydroxycinnamic acid (CHCA) matrix was added to each spot.
For Mycobacterium spp. isolates, 3 ml of liquid culture were transferred to a tube between 12–72 h after positivity. The tubes were centrifuged at 3000 g for 10 min, the supernatant was discarded, and the pellet was resuspended with 500 μl of 70% ethanol. Then, the suspension was transferred to a tube with glass beads and inactivated by vortexing for 15 min. After this step, the protein extraction procedure was the same as what was performed for Nocardia spp. isolates. Samples were analysed with the VITEK MS system (bioMérieux) with IVD (v.3.2) and RUO (v.4.17) modes. Spectra were acquired in the linear positive mode within the 2000–20,000 Da range. Escherichia coli strain (ATCC 8739) was used in each run for instrument calibration.
Identifications with values ≥99.0% and between 60.0% and 99.0% were considered as high and low‐confidence results, respectively. Values lower than 60.0% indicated two or three probable identifications. In these cases, concordance at genus or complex level was evaluated. When a disagreement was found, the identification obtained by DNA sequencing was considered as the reference.
Ethics approval
The Ethics Committee from the University Hospital Gregorio Marañon approved this study (Code: MALDI‐Micobacterias). The study was carried out using anonymized microbiological samples, not human products. Therefore, all the conditions to waive the informed consent have been met. The research procedures were conducted following the Declaration of Helsinki guidelines.
RESULTS
Identification of Nocardia isolates
The VITEK MS system yielded 84 (93.3%) isolates correctly identified at species level using the IVD mode and 77 (85.6%) isolates using the RUO mode (Table 1). With the RUO mode, 4 strains were correctly classified in comparison to reference spectra present in this database with a matching percentage lower than 60.0%: Nocardia asiatica (53.5%), Nocardia cyriacigeorgica (42.7%), Nocardia farcinica (53.2%) and Nocardia pneumoniae (54.3%). Regarding the misidentifications obtained, one isolate of N. pneumoniae was identified as Nocardia abscessus by the IVD mode with low‐confidence level (67.7%) and one isolate of Nocardia veterana was identified as Nocardia africana/nova by both IVD (99.9% confidence level) and RUO (84.0% confidence level) modes. In the two cases, the identification obtained belonged to a closely related species pertaining to the same complex (N. abscessus complex and N. nova complex, respectively). Considering this, a total of 95.6% and 86.7% of Nocardia isolates were correctly identified at species or complex level using IVD and RUO modes, respectively. Among the different species included in the study, Nocardia concava and N. pneumoniae are not included in VITEK MS IVD database, and thus, they could not be identified.
TABLE 1.
Identification obtained for Nocardia isolates by IVD and RUO modes
| Species | N | IVD | RUO | ||||
|---|---|---|---|---|---|---|---|
| Correct ID (%) | No ID (%) | Incorrect ID (%) | Correct ID (%) | No ID (%) | Incorrect ID (%) | ||
| N. abscessus | 14 | 14 (100) | 0 (0) | 0 (0) | 10 (71.4) | 4 (28.6) | 0 (0) |
| N. asiatica | 1 | 1 (100) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 0 (0) |
| N. brasiliensis | 1 | 1 (100) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 0 (0) |
| N. carnea | 1 | 1 (100) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 0 (0) |
| N. cyriacigeorgica | 34 | 34 (100) | 0 (0) | 0 (0) | 33 (97.1) | 1 (2.9) | 0 (0) |
| N. concava a | 1 | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 1 (100) | 0 (0) |
| N. farcinica | 17 | 17 (100) | 0 (0) | 0 (0) | 16 (94.1) | 1 (5.9) | 0 (0) |
| N. neocaledoniensis | 1 | 1 (100) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 0 (0) |
| N. nova | 4 | 4 (100) | 0 (0) | 0 (0) | 2 (50) | 2 (50) | 0 (0) |
| N. otitidiscavarium | 5 | 4 (80) | 1 (20) | 0 (0) | 4 (80) | 1 (20) | 0 (0) |
| N. pneumoniae a | 2 | 0 (0) | 1 (50) | 1 (50) b | 1 (50) | 1 (50) | 0 (0) |
| N. pseudobrasiliensis | 1 | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 1 (100) | 0 (0) |
| N. veterana | 6 | 5 (83.3) | 0 (0) | 1 (16.7) b | 5 (83.3) | 0 (0) | 1 (16.7) b |
| N. wallacei | 2 | 2 (100) | 0 (0) | 0 (0) | 2 (100) | 0 (0) | 0 (0) |
| Total | 90 | 84 (93.3) | 4 (4.4) | 2 (2.2) b | 77 (85.6) | 12 (13.3) | 1 (1.1) b |
Not included in VITEK MS database.
Identified as other close related species belonging to the same complex.
Identification of Mycobacterium isolates
Among Mycobacterium isolates, IVD mode identified correctly 96 (90.6%) isolates at species or complex level, two of them with confidence <99.0%: Mycobacterium fortuitum group (confidence level 45.8% and 25.0%). On the other hand, RUO mode identified 89 (84.0%) isolates at species or complex level (Table 2), three of them with values below 60.0%: Mycobacterium intracellulare (44.0%) and Mycobacterium kansasii (55.2% and 55.9%). Overall, no misidentifications were obtained by VITEK MS. Four strains of M. intracellulare were identified at genus level by RUO mode (confidence levels of 80.0%, 84.0%, 92.0% and 96.0%). In this study, 21 isolates belonging to M. fortuitum complex encompassing 4 species were included (M. fortuitum, Mycobacterium peregrinum, Mycobacterium porcinum and Mycobacterium septicum). The IVD mode identified all of them as M. fortuitum group, whereas RUO identified separately M. peregrinum and M. septicum. Three species analysed are not included in VITEK MS database, and they were not identified: Mycobacterium europaeum, Mycobacterium paragordonae and Mycobacterium rhodesiae.
TABLE 2.
Identification obtained for Mycobacterium isolates by IVD and RUO modes
| Species | N | IVD | RUO | ID genus level (%) | ||||
|---|---|---|---|---|---|---|---|---|
| Correct ID (%) | No ID (%) | Incorrect ID (%) | Correct ID (%) | No ID (%) | Incorrect ID (%) | |||
| M. abscessus | 18 | 18 (100) | 0 (0) | 0 (0) | 18 (100) | 0 (0) | 0 (0) | 0 (0) |
| M. avium | 12 | 11 (91.7) | 1 (8.3) | 0 (0) | 11 (91.7) | 1 (8.3) | 0 (0) | 0 (0) |
| M. chelonae | 1 | 1 (100) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 0 (0) |
| M. europaeum a | 3 | 0 (0) | 3 (100) | 0 (0) | 0 (0) | 3 (100) | 0 (0) | 0 (0) |
| M. fortuitum b | 13 | 13 (100) | 0 (0) | 0 (0) | 12 (92.3) | 1 (7.7) | 0 (0) | 0 (0) |
| M. gordonae | 10 | 9 (90) | 1 (10) | 0 (0) | 9 (90) | 1 (10) | 0 (0) | 0 (0) |
| M. intracellulare | 13 | 11 (84.6) | 2 (15.4) | 0 (0) | 9 (69.2) | 0 (0) | 0 (0) | 4 (30.8) |
| M. kansasii | 5 | 5 (100) | 0 (0) | 0 (0) | 5 (100) | 0 (0) | 0 (0) | 0 (0) |
| M. lentiflavum | 9 | 9 (100) | 0 (0) | 0 (0) | 7 (77.8) | 2 (22.2) | 0 (0) | 0 (0) |
| M. mageritense | 2 | 2 (100) | 0 (0) | 0 (0) | 1 (50) | 1 (50) | 0 (0) | 0 (0) |
| M. malmoense | 1 | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 0 (0) |
| M. paragordonae a | 1 | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 0 (0) |
| M. peregrinum | 3 | 3 (100) b | 0 (0) | 0 (0) | 3 (100) | 0 (0) | 0 (0) | 0 (0) |
| M. porcinum b | 4 | 4 (100) b | 0 (0) | 0 (0) | 4 (100) b | 0 (0) | 0 (0) | 0 (0) |
| M. rhodesiae a | 1 | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 0 (0) |
| M. septicum | 1 | 1 (100) b | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 0 (0) |
| M. shimoidei | 1 | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 0 (0) |
| M. smegmatis | 3 | 3 (100) | 0 (0) | 0 (0) | 3 (100) | 0 (0) | 0 (0) | 0 (0) |
| M. szulgai | 1 | 1 (100) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 0 (0) |
| M. triplex | 1 | 1 (100) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 0 (0) |
| M. xenopi | 3 | 3 (100) | 0 (0) | 0 (0) | 3 (100) | 0 (0) | 0 (0) | 0 (0) |
| Total | 106 | 96 (90.6) | 10 (9.4) | 0 (0) | 89 (84) | 13 (12.3) | 0 (0) | 4 (3.8) |
Not included in VITEK MS database.
Identified as M. fortuitum group.
DISCUSSION
The implementation of the VITEK MS system allowed 95.5% and 90.6% correct species or complex level identification of Nocardia spp. from plates and NTM isolates from liquid medium, respectively. In both cases, the IVD database provided a higher number of correct identifications. Only two Nocardia spp. isolates were misidentified at the species level and none of the NTM isolates (specificity of 97.7% for Nocardia and 100% for NTMs at the species level; specificity of 100% for both groups of microorganisms at the complex level).
In this study, the VITEK MS system coupled with an updated database (IVD v3.2) identified successfully 95.5% of Nocardia spp. isolates at species or complex level. By comparing both identification modes used, IVD and RUO, the latter reached a lower number of isolates identified. Previous studies using VITEK MS with IVD mode also showed high identification rates (87.0%–91.0%) for Nocardia spp. species (Body et al., 2018; Durand et al., 2020; Girard et al., 2016, 2017; Wei et al., 2019). Although different numbers of isolates and species were included in these studies, they showed similar results, demonstrating the robustness of the system for accurate identification of Nocardia strains.
Regarding misidentifications, only two isolates were misidentified at species level, although in both cases they belonged to the same complex. One of them was a N. pneumoniae isolate identified as N. abscessus (N. abscessus complex), due to the fact that N. pneumoniae is not included in IVD database but it is present in the RUO mode. The other misidentification consisted of a N. veterana isolate identified as N. africana/nova (N. nova complex); the same misidentification has also been observed in previous studies (Body et al., 2018; Girard et al., 2016).
For non‐tuberculous Mycobacterium spp. isolates, both modes combined (IVD and RUO) identified 86.7% of them, although the RUO identification rate was also lower than IVD. Other studies that have compared these two modes also obtained higher results using IVD mode, which could have relation with different identification algorithms used by them (Leyer et al., 2017; Wilen et al., 2015).
Of special interest is the fact that all isolates from the study have been analysed from a liquid culture medium. Previous studies focused on liquid media reported heterogeneous identification rates, from 69.4% to 98.8% (Huang et al., 2018; Kehrmann et al., 2016; Moreno et al., 2018; Park et al., 2016). These different results could be due to different protein extraction protocols performed, and different numbers and diversity of mycobacterial species analysed. In our study, similar results were obtained using IVD database v3.2, reaching 90.6% of isolates identified.
Interestingly, no incorrect species assignation was obtained, which shows the high reliability of this system for NTM identification. Only a few misidentifications have been reported in literature, involving close related species such as Mycobacterium avium and M. intracellulare (Miller et al., 2018), Mycobacterium immunogenum and M. abscessus (Brown‐Elliott et al., 2019) or Mycobacterium scrofulaceum and Mycobacterium parascrofulaceum (Luo et al., 2018). In the present study, although no M. scrofulaceum and M. immunogenum species were included, M. avium and M. intracellulare were correctly distinguished from each other. On the other hand, IVD mode was unable to differentiate members from M. fortuitum complex, as observed previously (Luo et al., 2018). However, RUO mode successfully identified M. peregrinum and M. septicum at the species level. This suggests that in the future, it could be possible to differentiate species from this complex by VITEK MS. Lastly, three of the species analysed (M. europaeum, M. paragordonae and M. rhodesiae) were not represented in the database and, as expected, they were unidentified.
In conclusion, the processing of Nocardia and Mycobacterium isolates with the VITEK MS Mycobacterium/Nocardia kit and their identification with the VITEK MS system demonstrated that this method is suitable for implementation in clinical microbiology laboratories for the robust, accurate and standardized identification of these microorganisms at the species level. Only 6.7% of the Nocardia isolates and 9.4% of the NTMs sent to the clinical microbiology laboratory for routine identification with VITEK MS may not to be reliably identified. In these cases, molecular methods must be applied to achieve a definitive identification. The limited number of isolates requiring confirmatory tests renders VITEK MS an efficient method for rapid identification of Nocardia and NTM isolates. Besides, the constant addition of less common species to its database will continue to benefit the identification accuracy obtained by this system and reduce the number or isolates requiring confirmatory testing.
AUTHOR CONTRIBUTIONS
DRT involved in formal analysis, data collection, validation, visualization, original draft preparation and review/editing. MEZ involved in formal analysis, supervision, validation, visualization, original draft preparation and review/ editing. PB, MBS, MM and MM and MJRS involved in data collection, validation and review/editing. PM involved in writing and review/editing. BRS involved in conceptualization, project administration, formal analysis, supervision, validation, visualization, original draft preparation and review/ editing.
FUNDING INFORMATION
This work was supported by the projects PI15/01073 and PI18/00997 from the Health Research Fund (FIS. Instituto de Salud Carlos III. Plan Nacional de I+D+I 2013–2016) of the Carlos III Health Institute (ISCIII, Madrid, Spain) partially financed by the European Regional Development Fund (FEDER) ‘A way of making Europe’. BRS (CPII19/00002) is recipient of a Miguel Servet contract supported by the FIS. DRT has been funded by the IiSGM through its intramural program.
CONFLICT OF INTEREST
BioMérieux SA provided a VITEK MS instrument for its evaluation as well as VITEK MS Mycobacterium/Nocardia kits for the preparation of isolates for MALDI‐TOF MS analysis.
Supporting information
Tables S1–S2
ACKNOWLEDGEMENT
This work was supported by the projects PI15/01073 and PI18/00997 from the Health Research Fund (FIS. Instituto de Salud Carlos III. Plan Nacional de I+D+I 2013–2016) of the Carlos III Health Institute (ISCIII, Madrid, Spain) partially financed by the European Regional Development Fund (FEDER) ‘A way of making Europe’. BRS (CPII19/00002) is recipient of a Miguel Servet contract supported by the FIS. DRT has been funded by the IiSGM through its intramural program.
Rodríguez‐Temporal, D. , Zvezdánova, M.E. , Benedí, P. , Marín, M. , Blázquez‐Sánchez, M. & Ruiz‐Serrano, M.J. et al. (2023) Identification of Nocardia and non‐tuberculous Mycobacterium species by MALDI‐TOF MS using the VITEK MS coupled to IVD and RUO databases. Microbial Biotechnology, 16, 778–783. Available from: 10.1111/1751-7915.14146
DATA AVAILABILITY STATEMENT
The protein spectra analysed in this project are available upon request.
REFERENCES
- Alcaide, F. , Pena, M.J. , Perez‐Risco, D. , Camprubi, D. , Gonzalez‐Luquero, L. , Grijota‐Camino, M.D. et al. (2017) Increasing isolation of rapidly growing mycobacteria in a low‐incidence setting of environmental mycobacteria, 1994‐2015. European Journal of Clinical Microbiology & Infectious Diseases, 36, 1425–1432. [DOI] [PubMed] [Google Scholar]
- Body, B.A. , Beard, M.A. , Slechta, E.S. , Hanson, K.E. , Barker, A.P. , Babady, N.E. et al. (2018) Evaluation of the Vitek MS v3.0 matrix‐assisted laser desorption ionization‐time of flight mass spectrometry system for identification of Mycobacterium and Nocardia species. Journal of Clinical Microbiology, 56, e00237‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown‐Elliott, B.A. , Fritsche, T.R. , Olson, B.J. , Vasireddy, S. , Vasireddy, R. , Iakhiaeva, E. et al. (2019) Comparison of two commercial matrix‐assisted laser desorption/ionization‐time of flight mass spectrometry (MALDI‐TOF MS) systems for identification of nontuberculous mycobacteria. American Journal of Clinical Pathology, 152, 527–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conville, P.S. , Brown‐Elliott, B.A. , Smith, T. & Zelazny, A.M. (2018) The complexities of Nocardia taxonomy and identification. Journal of Clinical Microbiology, 56, e01419‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand, T. , Vautrin, F. , Bergeron, E. , Girard, V. , Polsinelli, S. , Monnin, V. et al. (2020) Assessment of VITEK(R) MS IVD database V3.0 for identification of Nocardia spp. using two culture media and comparing direct smear and protein extraction procedures. European Journal of Clinical Microbiology & Infectious Diseases, 39, 559–567. [DOI] [PubMed] [Google Scholar]
- Falkinham, J.O., 3rd . (2016) Current epidemiologic trends of the nontuberculous mycobacteria (NTM). Current Environmental Health Reports, 3, 161–167. [DOI] [PubMed] [Google Scholar]
- Girard, V. , Mailler, S. , Welker, M. , Arsac, M. , Celliere, B. , Cotte‐Pattat, P.J. et al. (2016) Identification of Mycobacterium spp. and Nocardia spp. from solid and liquid cultures by matrix‐assisted laser desorption ionization‐time of flight mass spectrometry (MALDI‐TOF MS). Diagnostic Microbiology and Infectious Disease, 86, 277–283. [DOI] [PubMed] [Google Scholar]
- Girard, V. , Mailler, S. , Polsinelli, S. , Jacob, D. , Saccomani, M.C. , Celliere, B. et al. (2017) Routine identification of Nocardia species by MALDI‐TOF mass spectrometry. Diagnostic Microbiology and Infectious Disease, 87, 7–10. [DOI] [PubMed] [Google Scholar]
- Griffith, D.E. , Aksamit, T. , Brown‐Elliott, B.A. , Catanzaro, A. , Daley, C. , Gordin, F. et al. (2007) An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. American Journal of Respiratory and Critical Care Medicine, 175, 367–416. [DOI] [PubMed] [Google Scholar]
- Huang, T.S. , Lee, C.C. , Tu, H.Z. & Lee, S.S. (2018) Rapid identification of mycobacteria from positive MGIT broths of primary cultures by MALDI‐TOF mass spectrometry. PLoS One, 13, e0192291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrmann, J. , Schoerding, A.K. , Murali, R. , Wessel, S. , Koehling, H.L. , Mosel, F. et al. (2016) Performance of Vitek MS in identifying nontuberculous mycobacteria from MGIT liquid medium and Lowenstein‐Jensen solid medium. Diagnostic Microbiology and Infectious Disease, 84, 43–47. [DOI] [PubMed] [Google Scholar]
- Leyer, C. , Gregorowicz, G. , Mougari, F. , Raskine, L. , Cambau, E. & de Briel, D. (2017) Comparison of Saramis 4.12 and IVD 3.0 Vitek MS matrix‐assisted laser desorption ionization‐time of flight mass spectrometry for identification of mycobacteria from solid and liquid culture media. Journal of Clinical Microbiology, 55, 2045–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, L. , Cao, W. , Chen, W. , Zhang, R. , Jing, L. , Chen, H. et al. (2018) Evaluation of the VITEK MS knowledge base version 3.0 for the identification of clinically relevant Mycobacterium species. Emerging Microbes & Infections, 7, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makinen, J. , Marjamaki, M. , Marttila, H. & Soini, H. (2006) Evaluation of a novel strip test, genotype mycobacterium CM/AS, for species identification of mycobacterial cultures. Clinical Microbiology and Infection, 12, 481–483. [DOI] [PubMed] [Google Scholar]
- Marin, M. , Ruiz, A. , Iglesias, C. , Quiroga, L. , Cercenado, E. , Martin‐Rabadan, P. et al. (2018) Identification of Nocardia species from clinical isolates using MALDI‐TOF mass spectrometry. Clinical Microbiology and Infection, 24, 1342e1345–1342e1348. [DOI] [PubMed] [Google Scholar]
- Miller, E. , Cantrell, C. , Beard, M. , Derylak, A. , Babady, N.E. , McMillen, T. et al. (2018) Performance of Vitek MS v3.0 for identification of Mycobacterium species from patient samples by use of automated liquid medium systems. Journal of Clinical Microbiology, 56, e00219‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, E. , Miller, E. , Miller, E. , Totty, H. & Deol, P. (2018) A novel liquid media mycobacteria extraction method for MALDI‐TOF MS identification using VITEK(R) MS. Journal of Microbiological Methods, 144, 128–133. [DOI] [PubMed] [Google Scholar]
- Park, J.S. , Choi, S.H. , Hwang, S.M. , Hong, Y.J. , Kim, T.S. , Park, K.U. et al. (2016) The impact of protein extraction protocols on the performance of currently available MALDI‐TOF mass spectrometry for identification of mycobacterial clinical isolates cultured in liquid media. Clinica Chimica Acta, 460, 190–195. [DOI] [PubMed] [Google Scholar]
- Patel, R. (2015) MALDI‐TOF MS for the diagnosis of infectious diseases. Clinical Chemistry, 61, 100–111. [DOI] [PubMed] [Google Scholar]
- Rodriguez‐Sanchez, B. , Ruiz‐Serrano, M.J. , Ruiz, A. , Timke, M. , Kostrzewa, M. & Bouza, E. (2016) Evaluation of MALDI Biotyper Mycobacteria library v3.0 for identification of nontuberculous mycobacteria. Journal of Clinical Microbiology, 54, 1144–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez‐Temporal, D. , Perez‐Risco, D. , Struzka, E.A. , Mas, M. & Alcaide, F. (2017) Impact of updating the MALDI‐TOF MS database on the identification of nontuberculous mycobacteria. Journal of Mass Spectrometry, 52, 597–602. [DOI] [PubMed] [Google Scholar]
- Rodriguez‐Temporal, D. , Perez‐Risco, D. , Struzka, E.A. , Mas, M. & Alcaide, F. (2018) Evaluation of two protein extraction protocols based on freezing and mechanical disruption for identifying nontuberculous mycobacteria by matrix‐assisted laser desorption ionization‐time of flight mass spectrometry from liquid and solid cultures. Journal of Clinical Microbiology, 56, e01548‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez‐Temporal, D. , Alcaide, F. , Marekovic, I. , O'Connor, J.A. , Gorton, R. , van Ingen, J. et al. (2022) Multicentre study on the reproducibility of MALDI‐TOF MS for nontuberculous mycobacteria identification. Scientific Reports, 12, 1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleeb, P.G. , Drake, S.K. , Murray, P.R. & Zelazny, A.M. (2011) Identification of mycobacteria in solid‐culture media by matrix‐assisted laser desorption ionization‐time of flight mass spectrometry. Journal of Clinical Microbiology, 49, 1790–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinner, P. , Stenger, S. , Richter, E. , Brown‐Elliott, B. , Wallace, R. & Wengenack, N. (2015) Mycobacterium: laboratory characteristics of slowly growing mycobacteria. In: Manual of Clinical Microbiology. Washington, DC: ASM Press. 11th edition. [Google Scholar]
- Wei, M. , Wang, P. , Yang, C. & Gu, L. (2019) Molecular identification and phylogenetic relationships of clinical Nocardia isolates. Antonie Van Leeuwenhoek, 112, 1755–1766. [DOI] [PubMed] [Google Scholar]
- Wilen, C.B. , McMullen, A.R. & Burnham, C.A. (2015) Comparison of sample preparation methods, instrumentation platforms, and contemporary commercial databases for identification of clinically relevant mycobacteria by matrix‐assisted laser desorption ionization‐time of flight mass spectrometry. Journal of Clinical Microbiology, 53, 2308–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1–S2
Data Availability Statement
The protein spectra analysed in this project are available upon request.


