Fig. 2 |. Cis- versus trans-UCI conjugation.
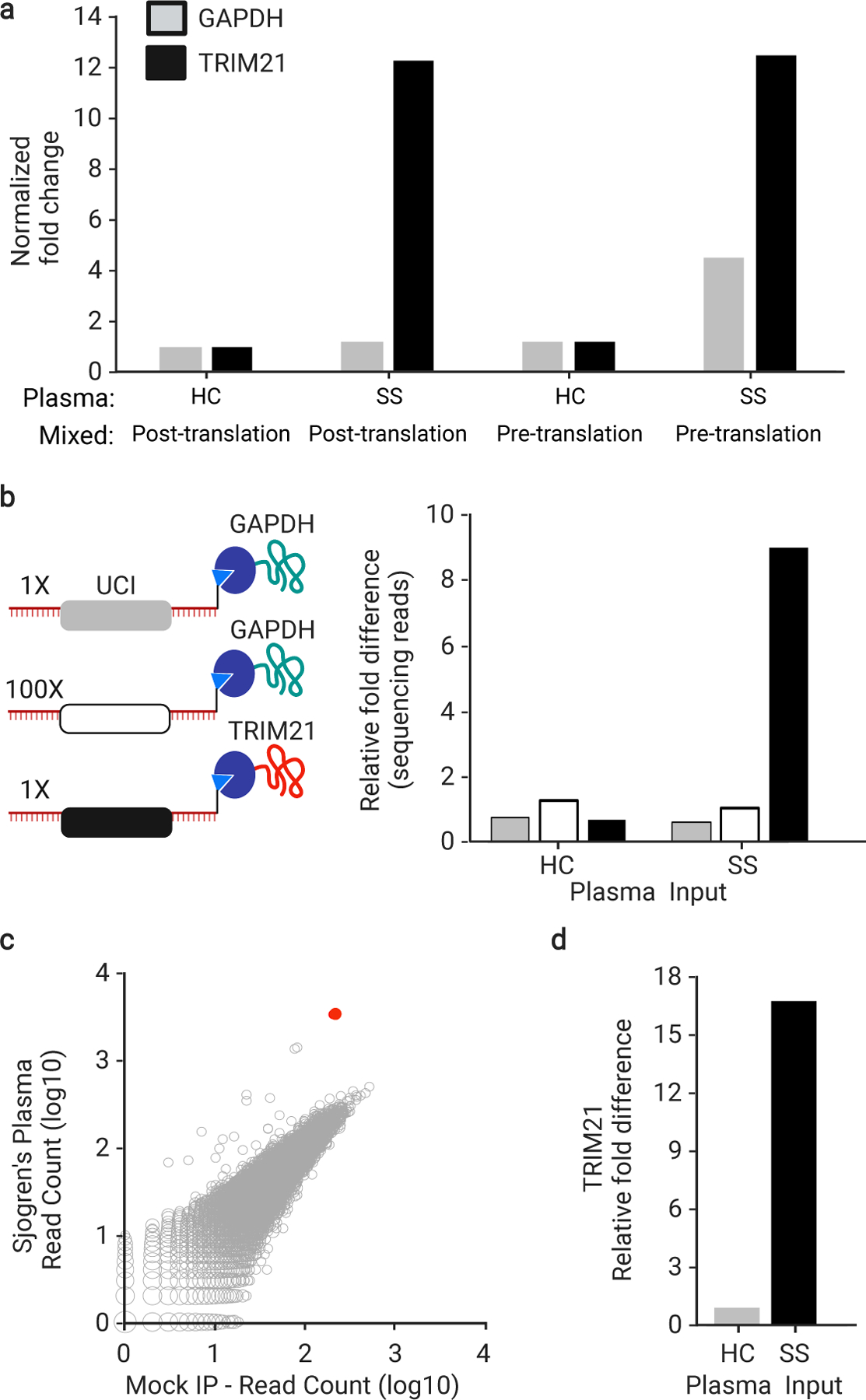
a, IVT-RNA encoding TRIM21 or GAPDH with their distinct UCI barcodes were translated before or after mixing at a 1:1 ratio. qPCR analysis of the IPs using UCI-specific primers, reported as fold-change versus IP with HC plasma, which is set to 1, when the IVT-RNA was mixed post-translation (technical replicates, n = 3). b, IVT-RNA encoding TRIM21 (black UCI) and GAPDH (gray UCI) were mixed 1:1 into a background of 100-fold excess GAPDH (white UCI) and then translated as a mock library. Sequencing analysis of the IPs, reported as fold-change versus the HC IP of the 100x GAPDH (technical replicates, n = 3). c, hORFeome MIPSA library containing spiked-in TRIM21, IPed with SS plasma and compared to average of 8 mock IPs (no plasma input). The TRIM21 UCI is shown in red. d, Relative fold difference of TRIM21 UCI in SS versus HC IPs, determined by sequencing.
