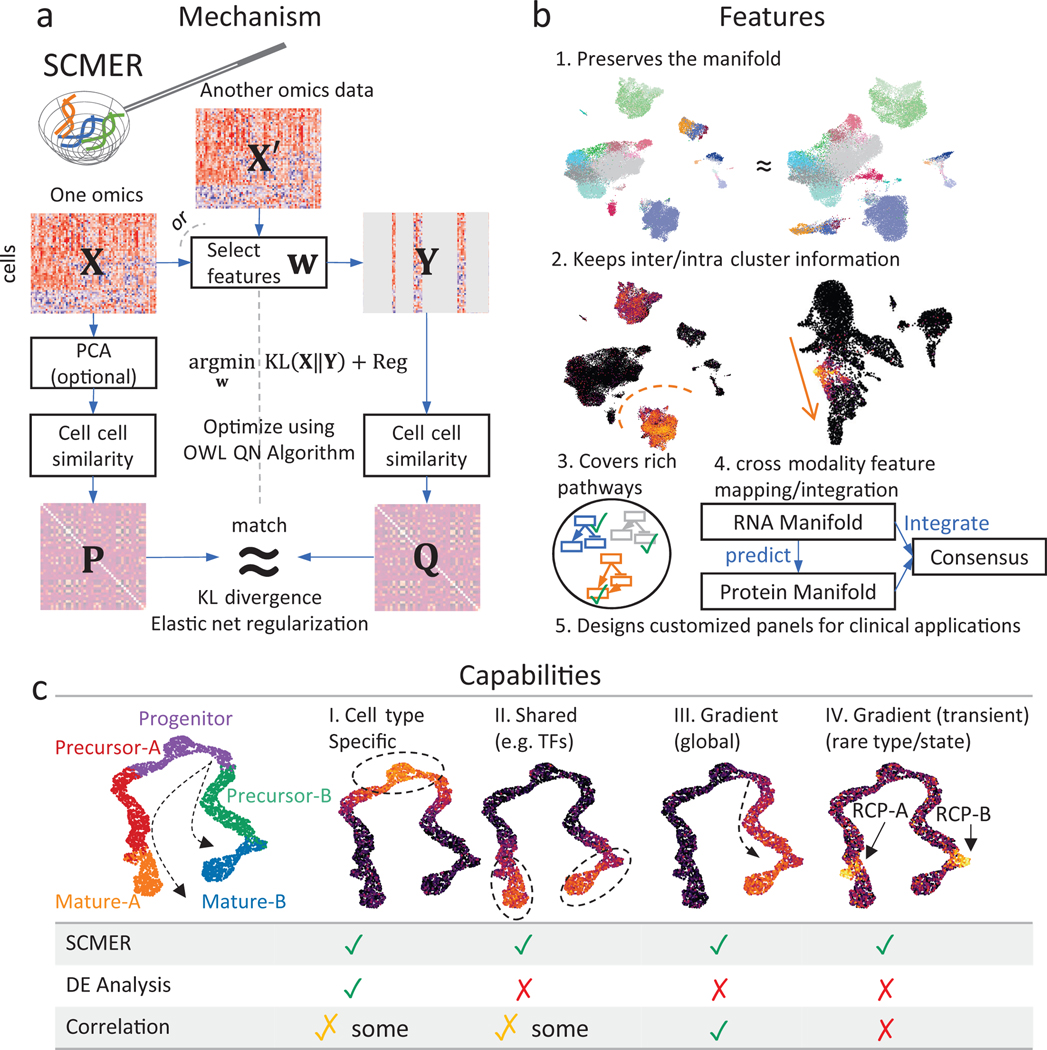
Fig. 1: The SCMER approach.
(a) Workflow of SCMER. SCMER selects the features that preserve the manifold from a single-cell omics dataset . Features can be selected from either or another co-assayed omics . Vector indicates the selection. is the dataset after feature selection. and are cell-cell similarity matrices for and , respectively.
(b) Applications of SCMER. SCMER selects features that preserve the manifold and retain inter- and intra-cluster diversity, and thus can be applied to discover rich molecular pathways, integrate modalities, and design customized DNA/RNA/antibody panels of restricted sizes.
(c) Capabilities of SCMER compared with mainstream label/cluster-based differential expression (DE) analysis methods and correlation-based methods. The hypothetical branching trajectories contain common progenitors, precursors for A and B, and mature A and B.