Extended Data Fig. 6 |. Multiple sclerosis SNPs within CD4 + T cell ATAC-Seq peaks.
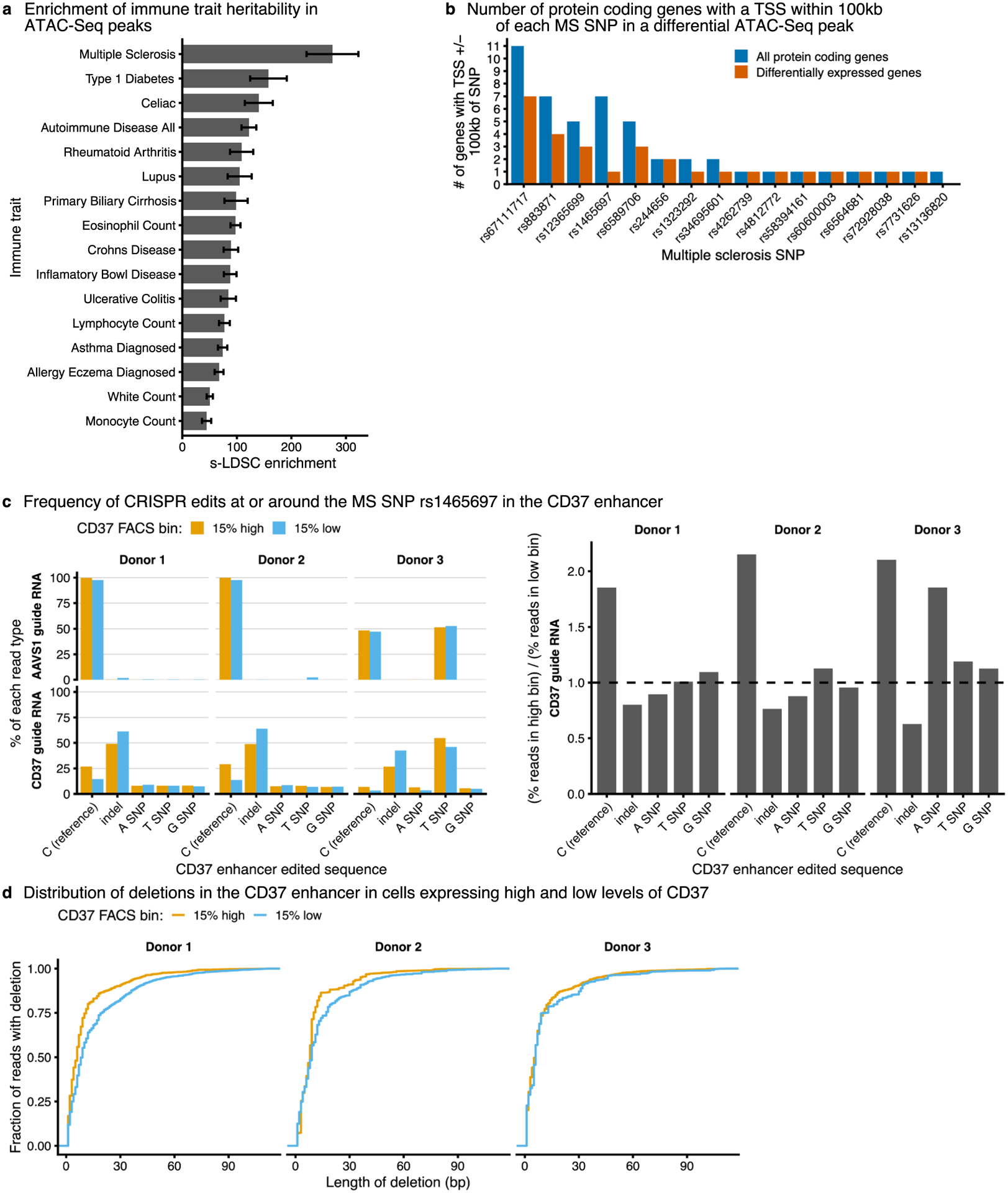
a, Enrichment of heritability in accessible ATAC-Seq peaks for different immune traits. Data are presented as estimated enrichment +/− standard error estimated from stratified LD score regression. b, The number of all protein-coding genes and differentially expressed protein-coding genes with a TSS within 100 kb of a multiple sclerosis SNP. Only high confidence multiple sclerosis SNPs (PICS probability greater than 50%) within differentially accessible ATAC-Seq peaks are shown. c, Editing outcomes in CD37 low and high-expressing cells after using CRISPR/Cas9 and homology-directed repair templates to edit the SNP rs1465697. Editing was performed with guide RNAs targeting the CD37 CRE (CD37 guide RNA) or a control region (AAVS1 guide RNA). d, Length of deletions after CRISPR editing in CD37 low- and high-expressing cells.
