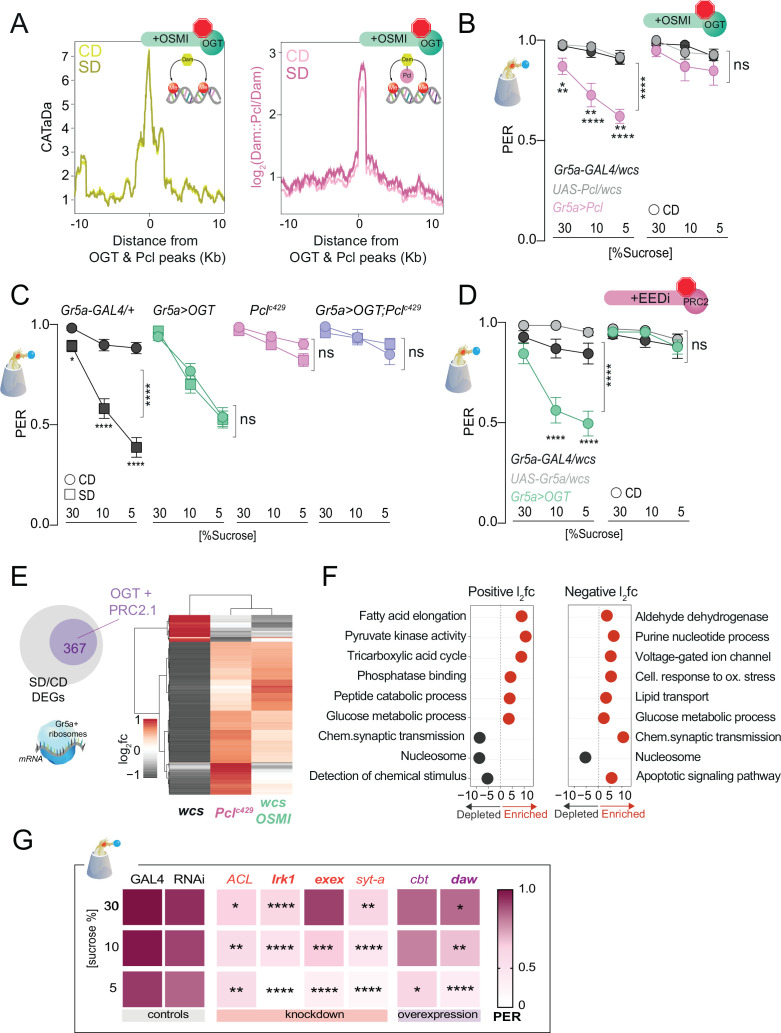
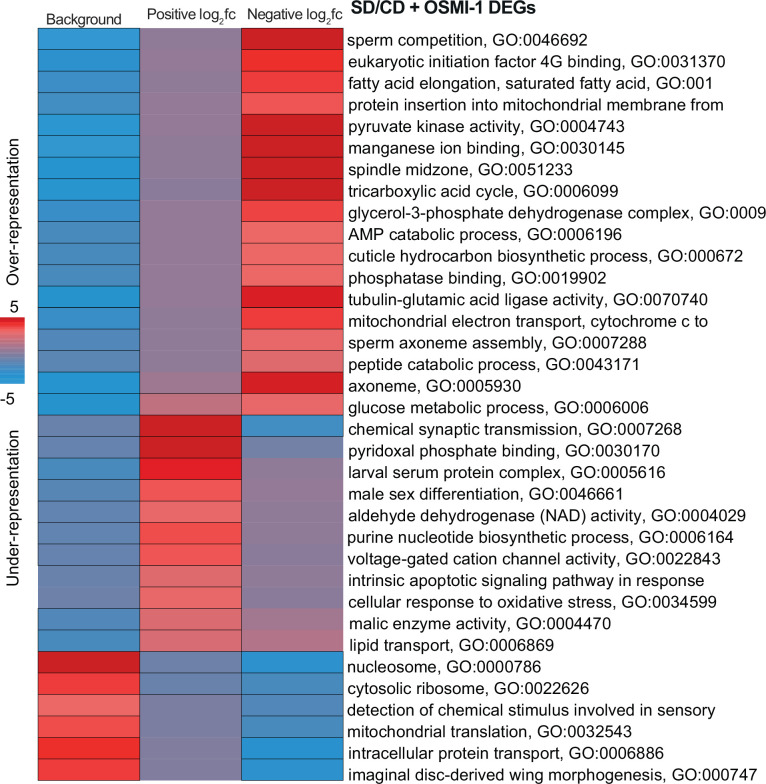
Figure 4. O-GlcNAc Transferase (OGT) activity is necessary for chromatin and transcriptional dynamics in response to the sugar diet environment.
(A) (left) Average CATaDa signal on a CD (light yellow) and SD (dark yellow) with OSMI centered at OGT + Pcl peaks (compare to Figure 2D); (right) Average log2 Dam::Pcl/Dam signal on a CD (light pink) and SD (dark pink) with OSMI centered at OGT + Pcl peaks (compare to Figure 2). (B) Taste responses (y-axis) to stimulation of the labellum with 30, 10, and 5% sucrose (x-axis) of age-matched male Gr5a>Pcl (pink) and transgenic controls (shades of gray) on CD supplemented with vehicle (DMSO, n = 14–23) or 10 µM OSMI (n = 20–42). Two-way repeated measure analysis of variance (ANOVA): (1) DMSO, main effect of genotype (****p < 0.0001) and genotype x concentration (*p < 0.05); Tukey multiple comparisons test for 30, 10, and 5% sucrose concentrations: Gr5a>wcs vs. Gr5a>Pcl p = 0.0165, p = 0.0056, p = 0.0025; Pcl>wcs vs. Gr5a>wcs, ns. (2) OSMI: main effect of genotype p = 0.3194 and genotype × concentration p = 0.6893. (C) Taste responses (y-axis) to stimulation of the labellum with 30, 10, and 5% sucrose (x-axis) in Gr5a>OGT;Pclc429 (purple), Gr5a>OGT (green), Pclc429/+ (pink), and transgenic controls (gray) on CD or SD, n = 20–37. Two way repeated measure ANOVA, main effect of diet: ****p < 0.0001. Tukey multiple comparisons test, ****p < 0.0001 and *p < 0.05. (D) Taste responses (y-axis) to stimulation of the labellum with 30, 10, and 5% sucrose (x-axis) in Gr5a>OGT (green) and transgenic controls (shades of gray) on a CD supplemented with vehicle or 8 µM EEDi, n = 20–22. Two-way repeated measure ANOVA, main effect of Gr5a>OGT genotype compared to each control genotype: ****p < 0.0001. Tukey multiple comparisons test, ****p < 0.0001. (E) Log2fold (l2fc) of differentially expressed genes (DEGs) between SD/CD in w1118cs ± OSMI and Pclc429 SD/CD. (F) GO term analysis of the DEGs measured in the Gr5a+ neurons of flies fed a CD and SD + OSMI. (G) Taste responses (y-axis) to stimulation of the labellum with 30, 10, and 5% sucrose (x-axis) for a subset of DEGs in (E, purple circle) that show dependence on OGT and Polycomb Repressive Complex 2.1 (PRC2.1). n = 14–49. Purple, knockdown; red, overexpression; bold, direct OGT/PRC2.1 targets. Two-way repeated measure ANOVA, main effect of GAL4>wcs control genotype compared to each control genotype: ****p <0.0001. Tukey multiple comparisons test, ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05. ACL, ATP Citrate Lyase; Irk1, inwardly rectifier potassium channel 1; daw, dawdle; cbt, cabut; exex, extra extra; syt-a, synaptotagmin alpha. Data are shown as mean ± standard error of the mean (SEM).