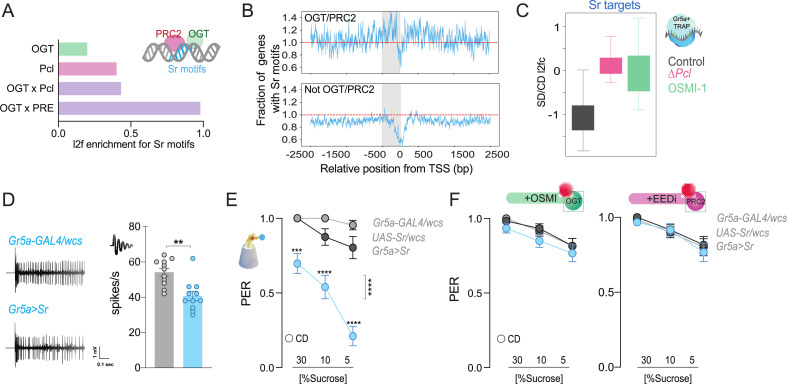
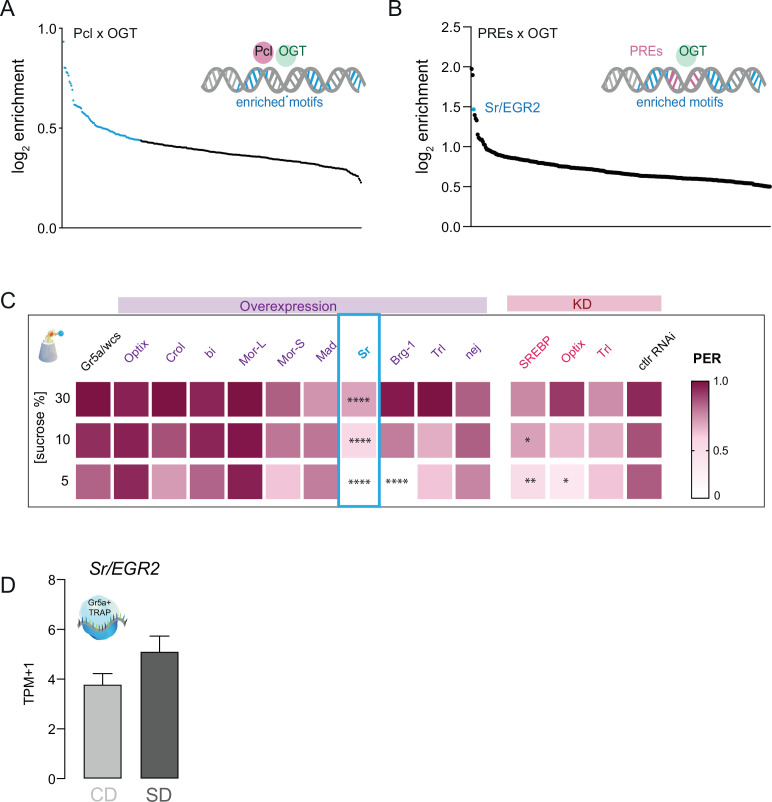
Figure 5. The immediate early gene Sr is found at PRC2 × OGT genes and is involved in sweet-taste sensation.
(A) Log2fold (l2f) enrichment for Sr motifs at sites occupied by O-GlcNAc Transferase (OGT; green), Pcl (pink), or OGT + Pcl and OGT + PREs (purple), p < 0.0001. (B) Normalized distribution of Sr motifs along the regulatory regions 2500 bp up and downstream the transcriptional start site (TSS) for the genes in (A); the 500 bp immediately before each TSS are shaded in grey. Counts are normalized relative to the genome-wide expectation (derived by multiplying the number of potential target genes by the fractional coverage of Sr motifs on the genome); a score of 1.0 indicates the hypothetical genome-wide average overlap with Sr motifs and is shown as a red dashed line. (C) The distribution of RNA l2fc for genes that have Sr sites and are expressed in the Gr5a+ neurons of flies on a CD and SD in control flies or flies with mutations in Pcl or inhibition of OGT. q > 0.01. (D) Representative traces (left) and averaged neuronal responses to 25 mM sucrose stimulation from the L-type sensilla of Sr overexpression flies (blue) and controls (gray). n = 11. Mann–Whitney test: **p = 0.001. (E) Taste responses (y-axis) to stimulation of the labellum with 30, 10, and 5% sucrose (x-axis) in Sr overexpression flies (blue) and controls (gray). n = 22–38. Two-way repeated measure analysis of variance (ANOVA), main effect of genotype ****p < 0.0001 and genotype × concentration ****p < 0.0001; Tukey post-test for multiple comparisons: 30%: ***p = 0.0002 for Gr5a>Sr compared to each control, 10%: Gr5a>Sr vs. Gr5a>wcs p = 0.0025 and Gr5a>Sr vs. Sr >wcs ****p < 0.0001; 5%: ****p < 0.0001 for Gr5a>Sr compared to each control. Gr5a>wcs vs. Sr>wcs p > 0.05 at all concentrations. (F) Taste responses (y-axis) to stimulation of the labellum with 30, 10, and 5% sucrose (x-axis) in Sr overexpression flies (blue) and controls (gray) treated with the OGT inhibitor OSMI (green) or the PRC2 inhibitor EEDi (pink). n = 30. Two-way repeated measure ANOVA, main effect of genotype p = 0.2993 and p = 0.9146 and genotype × concentration p = 0.9293 and p = 0.9146, respectively. Data are shown as mean ± standard error of the mean (SEM).