Figure 1.
Enhanced CNS tropism of systemically administered BR1N
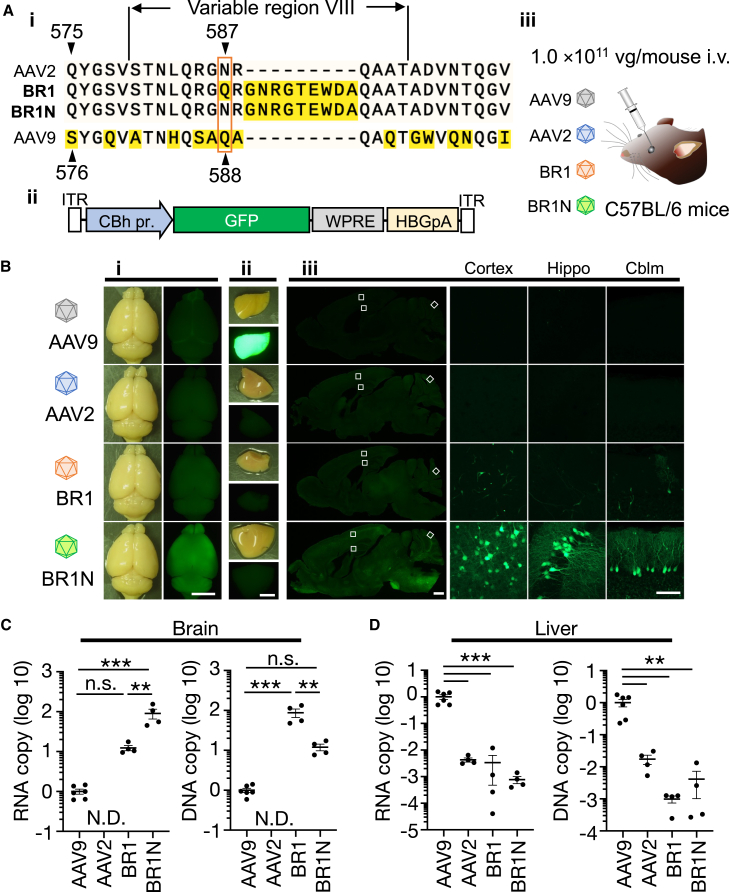
(A-i) Amino acid sequence alignment of AAV capsids (AAV2, BR1, BR1N, and AAV9) around variable region VIII. BR1 and BR1N, AAV2 capsid mutants, have a 7-amino-acid (NRGTEWD) insertion with a “stuffer” sequence (G and A) at both sides in the AAV2 capsid between positions 588 and 589. BR1 also has an asparagine (N)-to-glutamine (Q) substitution at position 587. Amino acids distinct from those of AAV2 are highlighted by a yellow background. (A-ii) Schematic depicting the AAV vector genome. GFP is expressed under control of the CBh promoter. ITR, inverted terminal repeat; WPRE, woodchuck hepatitis virus post-transcriptional regulatory element; HBGpA, human β-globin poly(A). (A-iii) AAV9, AAV2, BR1, or BR1N (1 × 1011 vg/mouse, respectively) was injected into adult C57BL/6 mice intravenously (i.v.) through the orbital venous plexus. (B) GFP fluorescence and bright-field images of whole brains (B-i), livers (B-ii), and sagittal brain sections (B-iii) 3 weeks after the AAV injection. Panels in the first to third rows from the right show magnifications of the squares in the cerebral cortex (cortex), hippocampus (Hippo), and cerebellar cortex (Cblm) in the low-power sagittal sections. Scale bars: 5 mm (i and ii), 1 mm (left in iii), and 100 μm (right in iii). (C and D) Logarithmic graphs showing amounts of AAV-derived mRNA and AAV genome. Cortex and liver tissues from female mice treated with AAV9, AAV2, BR1, or BR1N were examined 3 weeks after virus injection. Asterisks indicate statistically significant differences (n = 4–6 mice per group; ∗∗p < 0.01, ∗∗∗p < 0.001 by one-way ANOVA with Bonferroni’s post hoc test for C and D); N.D., not detected; n.s., not significant). All error bars show SEM.