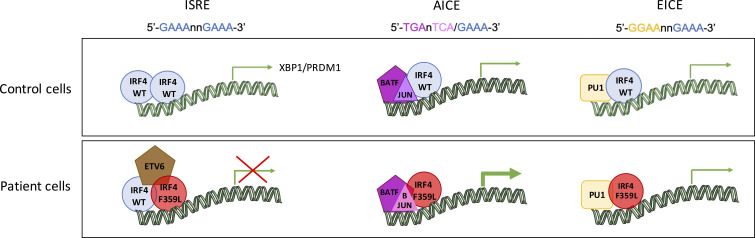
Thouenon et al. describe a novel primary immunodeficiency associated with defective plasma cell differentiation. They reveal that an amino acid exchange located within the interferon activation domain of IRF4 downregulates transcription on ISRE sites because of altered protein interaction.
Abstract
Here, we report on a heterozygous interferon regulatory factor 4 (IRF4) missense variant identified in three patients from a multigeneration family with hypogammaglobulinemia. Patients’ low blood plasmablast/plasma cell and naïve CD4 and CD8 T cell counts contrasted with high terminal effector CD4 and CD8 T cell counts. Expression of the mutant IRF4 protein in control lymphoblastoid B cell lines reduced the expression of BLIMP-1 and XBP1 (key transcription factors in plasma cell differentiation). In B cell lines, the mutant IRF4 protein as wildtype was found to bind to known IRF4 binding motifs. The mutant IRF4 failed to efficiently regulate the transcriptional activity of interferon-stimulated response elements (ISREs). Rapid immunoprecipitation mass spectrometry of endogenous proteins indicated that the mutant and wildtype IRF4 proteins differed with regard to their respective sets of binding partners. Our findings highlight a novel mechanism for autosomal-dominant primary immunodeficiency through altered protein binding by mutant IRF4 at ISRE, leading to defective plasma cell differentiation.
Graphical Abstract
Introduction
Characterization of the pathophysiological mechanisms that underlie newly recognized primary immunodeficiencies (PIDs) provides researchers with a unique opportunity to study the molecular details of the human immune system. The most common PIDs in humans are primary antibody deficiencies (PADs), which can result from intrinsic or extrinsic defects in B cell development, antibody maturation, plasma cell differentiation, and/or T cell development (Durandy et al., 2013). Although the genetic diagnosis of PAD patients is improving rapidly (due notably to greater availability of next-generation sequencing), most affected individuals still do not obtain a firm molecular diagnosis (Bousfiha et al., 2020; Fusaro et al., 2021; Tangye et al., 2020).
Interferon regulatory factor 4 (IRF4, also known as NF-EM5, Pip, LSIRF, ICSAT, and MUM1) belongs to a family of nine human transcription factors (De Silva et al., 2012). Unlike the other eight family members, IRF4 is not regulated by interferons (Nam and Lim, 2016). IRF4 expression is activated by antigen receptor signaling, TLRs, and CD40 (De Silva et al., 2012; Negishi et al., 2005). Initial studies of murine models demonstrated the IRF4’s essential role in IgM, IgG, and IgA secretion, antibody responses, and the generation of germinal center B cells (Mittrücker et al., 1997). Later studies revealed that IRF4’s function is required for Ig class-switch recombination and plasma cell differentiation (Klein et al., 2006; Sciammas et al., 2006). The differentiation of activated B lymphocytes into plasma cells depends on IRF4-induced expression of the PRDM1 gene (coding for BLIMP-1; Klein et al., 2006; Sciammas et al., 2006). In the “kinetic control” model, signaling-induced changes in the IRF4 protein level control the fate of activated B lymphocytes (Ochiai et al., 2013). According to this model, the IRF4 gene locus “senses” the strength of antigen receptor signaling, and the IRF4 protein “writes” the B cells’ trajectories. It has been shown that low levels of IRF4 expression favor the differentiation of BCL6-expressing germinal center B cells, whereas high levels favor the expression of BLIMP-1–expressing plasma cells (Ochiai et al., 2013). Along with this important role in B cell–mediated immunity, IRF4’s function is also involved in the differentiation and/or function of several T cell populations, including CD4+ T helper 2 (Th2), Th9, Th17, and T follicular helper (TFH) cells, effector regulatory T cells, and CD8+ cytotoxic effector and memory T cells (Huber and Lohoff, 2014). As with the model suggested for B lymphocyte fate, it has been suggested that IRF4 senses the TCR signaling strength (Krishnamoorthy et al., 2017) and thus acts as a rheostat by translating TCR avidity into appropriate transcriptional programs (Man et al., 2013). IRF4’s ability to control various transcriptional programs arises from its interactions with several transcriptional partners (Remesh et al., 2015). The IRF4 protein (like all members of the IRF family) contains two conserved functional domains: an N-terminal helix-turn-helix DNA-binding domain (DBD) containing conserved tryptophan residues and a C-terminal interferon activation domain (IAD, also known as the IRF-association domain) known to be critical in mediating protein–protein interactions (Remesh et al., 2015; Sundararaj et al., 2021). In contrast to the other IRF family members, the IRF4 protein contains a C-terminal flexible autoinhibitory region that binds directly to the DBD and modulates the interaction with target DNA (Remesh et al., 2015). Various DNA binding motifs have been described for IRF4 depending on the interaction partner and the cellular context. High IRF4 concentration within cells enables IRF4’s binding to the canonical interferon-stimulated response element (ISRE), allowing to establish IRF4 homodimer DNA complexes (Sundararaj et al., 2021). These stable ternary complexes are known to have an important role in plasma cell differentiation and BLIMP-1 expression (Sciammas et al., 2006). As a heterodimer with the PU.1, SPIB, or BATF transcription factors, IRF4 binds to erythroblast-transformation-specific interferon composite elements (EICEs) or AP-1-IRF composite elements (AICE 1 or 2), respectively, and thus mediates transcriptional activity (Brass et al., 1996; Li et al., 2012; Ochiai et al., 2013). The interactions between erythroblast-transformation-specific transcription factors and IRF4 have been described especially in B lymphocytes and dendritic cells, whereas the functions of the heterodimeric complexes between AP-1 and IRF4 have been described for T and B lymphocytes (Brass et al., 1996; Li et al., 2012; Ochiai et al., 2013). Recently, it was reported that a complex including IKAROS and IRF4 binds to zinc finger-IRF composite elements and represses a subset of genes during plasma cell differentiation (Ochiai et al., 2018).
It has also been reported that IRF4 deficiency (associated with a splice acceptor site mutation [c.1213-2A>G,pV405GfsTer127; NM_001195286] in the IRF4 gene, in conjunction with uniparental isodisomy) causes a PID (Bravo García-Morato et al., 2018). This patient suffered from a combined immunodeficiency with agammaglobulinemia, eosinophilia, normal lymphocyte counts, low memory T and B cell counts, and elevated GM-CSF–induced macrophage polarization (Bravo García-Morato et al., 2018). Age-dependent, incomplete penetrance of Whipple’s disease (a clinical manifestation due to Tropheryma whipplei [Tw] exposure) has been associated with IRF4 haploinsufficiency caused by a heterozygous loss-of-function mutation affecting IRF4’s DBD (Guérin et al., 2018). A genome-wide association study found that a single nucleotide polymorphism (SNP; rs12203592, located in intron 4 of the IRF4 gene) was linked to hair and skin color, indicating that IRF4 also has a role outside the immune system (Han et al., 2008). Subsequent functional studies demonstrated that this SNP lies within an enhancer of IRF4 transcription in melanocytes and impairs the binding of the TFAP2A transcription factor that (together with the melanocyte master regulator MITF) regulates the enhancer’s activity (Praetorius et al., 2013). Another genome-wide association study linked this SNP to not only hair color but also hair graying (Adhikari et al., 2016).
Here, based on our investigation of a multigeneration family, we describe a novel autosomal dominant PAD caused by a pathogenic IRF4 variant affecting the IAD. All three patients in the family presented with low IgM, IgG, and IgA serum levels (diagnosed during childhood); low plasma cell counts; abnormal T cell subsets; and early hair graying.
Results
Clinical history
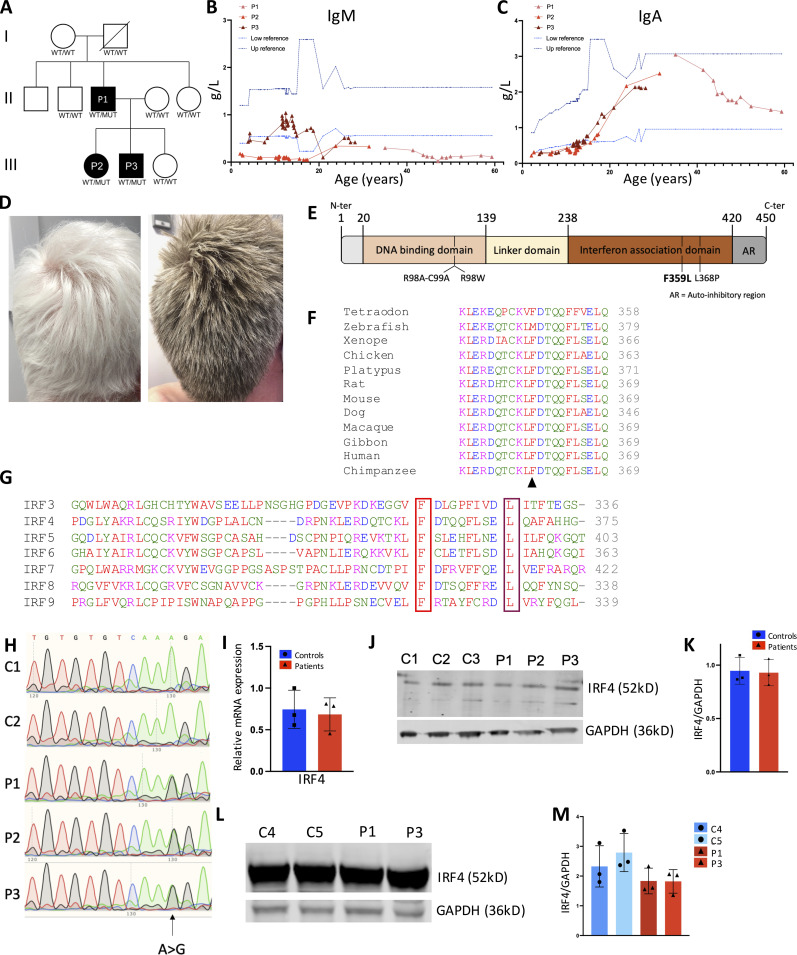
We report herein on three patients from two generations of a family affected by a PAD (Fig. 1 A). Both of the male index patient’s (P1) parents and the patient’s two brothers and sister were reportedly healthy. Patient P1 started to suffer from diarrhea at 6 mo of age. Due to recurrent nasopharyngitis and persistence of diarrhea at the age of 11 mo, P1 was diagnosed with panhypogammaglobulinemia (reported as affecting IgM, IgG, and IgA, although the levels were not available) and put on a gluten-free diet for 6 yr. At the age of 15 mo, he was hospitalized for severe diarrhea and Ig replacement therapy was initiated. He also suffered an episode of pneumonitis at the age of 28 mo. His tonsils and adenoids were removed at 7.5 yr of age. From the age of 8 until his last evaluation in adulthood (at the age of 60), P1 had normal IgA levels and low IgM levels. P1 experienced a meningococcal infection (at the age of 15 yr), a Giardia lamblia infection, a rectal cytomegalovirus infection, two separate disseminated varicella zoster virus infections, onychomycosis, and oral mycosis (all in adulthood). The persistence of diarrhea (it is noteworthy that the patient was negative for Tw infection in a PCR test) led to the diagnosis of inflammatory bowel disease and initiation of an efficient treatment with an antitumor necrosis factor agent when the patient was 48 yr of age. At the age of 56, P1 presented with hepatosplenomegaly. A liver biopsy indicated (predominant CD8) T cell infiltration. Nonhomogeneous hair pigmentation changes were reported from 12 yr of age onward and evolved into a gray color 2 yr later; at 25 yr of age, P1 had a completely gray head of hair. Skin lesions with depigmentation spots (diameter: 3 cm) had been also noted (Fig. S1 A).
Figure 1.
A novel hypogammaglobulinemia caused by a heterozygous IRF4 mutation. (A) The pedigree of the index family with allele segregation. (B and C) Changes in serum IgM (B) and IgA (C) levels over time for the patients (P1: pink; P2: light red; P3: dark red). The lines indicate the upper and lower reference boundaries. L, liter. (D) The hair of P1 and P3 (at the ages of 60 and 26, respectively). (E) Schematic representation of the IRF4 protein’s functional domains, depicting the positions of loss of DNA binding mutations (R98A-C99A and R98W), a loss of interaction with PU-1 variant (L368P), and the novel F359L variant. (F) Alignment of human IRF4 with orthologs. Protein sequences were extracted from the Ensembl Genome Browser and aligned using Clustal Omega. The black arrowhead indicates the location of the patient’s mutation (F359). (G) Alignment of IRF4’s IAD (around the F359L [in red] and L368P [in violet] variants) with IRF family members IRF3-IRF9. (H) Sanger sequencing of RT-PCR products of T cells blast generated from two controls and all the patients. (I) qPCR analysis of IRF4 transcripts from T cells blast generated from controls and patients. Data represent mean of n = 3 controls and n = 3 patients ± SD. (J–M) IRF4 expression levels in T cell blasts (J and K) or B-EBV cells (L and M) were analyzed in Western blots of total cell lysates from control (C) and patient (P). GAPDH was used as loading control. (K) Data of quantification represent mean of n = 3 controls and n = 3 patients ± SD. (L) Representative images and (M) data represent mean ± SD of quantifications of n = 3 repetition with n = 2 patient and n = 2 control derived B-EBV cells.
Figure S1.
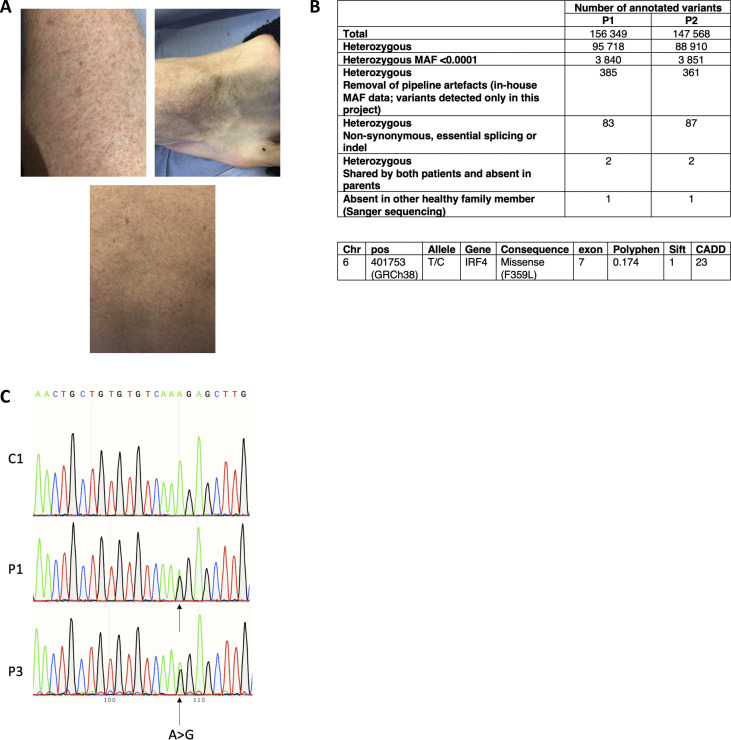
Clinical features and genetic analysis. (A) Skin manifestations observed for P3 at 26 yr of age. (B) A table summarizing the different filters applied to whole-exome sequencing data from patients to identify possible disease-causing variants. The analysis was performed for P1, P3, and P1’s parents and sister. A strict de novo model involved P1 and his parents only. The two variants identified in the strict de novo model are annotated. (C) Sanger sequencing of RT-PCR products of B-EBV cells derived from n = 1 control and n = 2 patients (P1 and P3).
P1’s daughter (P2) and son (P3) presented with recurrent ear, nose, and throat infections from early childhood onward. P2 and P3 were screened for PIDs at the age of 4 and 2 yr, respectively; panhypogammaglobulinemia was observed (Fig. 1, B and C, and Table 1) and Ig replacement therapy was initiated in both cases. The IgA levels (and IgM levels, for P3) normalized over time. P2’s clinical manifestations also included conjunctivitis, severe varicella, recurrent herpes virus infections, and a systemic Bartonella henselae infection at 9 yr of age. P3 had his tonsils and adenoids removed at 4 yr of age, and a molluscum contagiosum virus infection was reported during childhood. In adulthood, P3 experienced onychomycosis and cutaneous infection due to Epidermophyton floccosum, and diarrhea. P2 and P3 both presented with early hair graying as their father and P3 with the same skin depigmentation lesions (Fig. 1 D and Fig. S1 A).
Table 1.
Immunophenotyping results from the hospital laboratory
| P1 | P2 | P3 | Standards | |||||
|---|---|---|---|---|---|---|---|---|
| Sex | Male | Female | Male | |||||
| Age (at initial evaluation; mo) | 11 | 48 | 24 | |||||
| Serum IgG levels (g/liter) | NA | 2.27 | 2.50 | 7–14 | ||||
| Serum IgA levels (g/liter) | NA | 0.26 | 0.22 | 0.90–4 | ||||
| Serum IgM levels (g/liter) | NA | 0.43 | 0.18 | 0.50–2.50 | ||||
| Age (at evaluation; yr) | 35 | 31 | 28 | |||||
| Serum IgE levels (kUI/liter) | <2 | <5 | <5 | <100 | ||||
| P1 | P2 | P3 | Standards | |||||
| Age (at evaluation; yr) | 53 | 57 | 60 | 31 | 21 | 26 | 28 | |
| T lymphocytes | ||||||||
| CD3+ (/µl) | 590 | 344 | 819 | 903 | 4,421 | 1,157 | 1,928 | 1,008–1,647 |
| CD3+ CD4+ (/µl) | 181 | 116 | 284 | 457 | 1,350 | 450 | 597 | 480–1,320 |
| Activated CD4+ (HLA-DR+/CD3+CD4+; %) | 18.3 | NA | NA | NA | 12 | 11.1 | NA | 5–12 |
| Naïve CD4+ (CD45RA+CCR7+/CD3+CD4+; %) | 7.5 | 8.8 | 4.6 | 11.9 | 10.7 | 15.0 | 10.8 | 26–54 |
| Memory CD4+ (CD45ROb+/CD3+CD4+; %) | 84.7 | NA | NA | NA | 54.5 | 72.7 | NA | 40–61 |
| Central memory CD4+ (CD45RA−CCR7+/CD3+CD4+; %) | 43.8 | 59.2 | 64.2 | 57.4 | 20 | 35.5 | 24.3 | 28–51 |
| Mem effector CD4+ (CD45RA−CCR7−/CD3+CD4+; %) | 38.5 | 25 | 26.3 | 18.2 | 32.5 | 39.5 | 42.8 | 7.8–23.2 |
| Terminal effector CD4+ (CD45RA+CCR7−/CD3+CD4+; %) | 10.3 | 6.9 | 4.9 | 12.6 | 36.8 | 10.1 | 22.2 | 0–2.9 |
| CD3+ CD8+ (/µl) | 404 | 225 | 539 | 446 | 3,074 | 691 | 1,290 | 192–720 |
| Activated CD8+ (HLA-DR+/CD3+CD8+; %) | 32.8 | NA | NA | NA | 19.1 | 19.6 | NA | 9–33 |
| Naïve CD8+ (CD45RA+CCR7+/CD3+CD8+; %) | 1.2 | 1.7 | 1.9 | 5.7 | 3.0 | 6.7 | 3.3 | 24.2–53.6 |
| Memory CD8+ (CD45ROb+/CD3+CD8+; %) | 54.6 | NA | NA | NA | 63.4 | 73.9 | NA | 21–48 |
| Central memory CD8+ (CD45RA−CCR7+/CD3+CD8+; %) | 4.3 | 4.9 | 7.7 | 6.5 | 1.2 | 3.8 | 2.6 | 5.1–19.6 |
| Mem effector CD8+ (CD45RA−CCR7−/CD3+CD8+; %) | 37.7 | 34.1 | 39.1 | 30.1 | 58.6 | 73.0 | 73.5 | 16.4–32.6 |
| Terminal effector CD8+ (CD45RA+CCR7−/CD3+CD8+; %) | 56.8 | 59.4 | 51.3 | 57.8 | 37.3 | 16.5 | 20.5 | 5.2–37.4 |
| NK cells | ||||||||
| CD3−CD16+CD56+ (/µl) | 19 | 18 | 44 | 94 | NA | 82 | 69 | 56–400 |
| B lymphocytes | ||||||||
| CD19+ (/µl) | 260 | 30 | 83 | 84 | 239 | 123 | 127 | 67–270 |
| Naïve B cells (CD27−IgD+/CD19+; %) | 88 | 85.2 | 70.7 | 64.2 | 41.4 | 42.6 | 50.8 | 51–77 |
| Memory B cells (CD27+/CD19+; %) | 12 | 10.9 | 16.7 | 32.3 | 52 | 49.3 | 41.1 | 23–49 |
| Un-switched mem (CD27+IgD+/CD19+; %) | 6.4 | 5.1 | 9.5 | 15.4 | 45.5 | 32.5 | 31.3 | 3–40 |
| Switched mem (CD27+IgD−/CD19+; %) | 5 | 6.1 | 6.8 | 16.5 | 7.7 | 16.6 | 11.1 | 4.4–20.5 |
| CD21low B cells (CD19+CD21low; %) | 2.5 | 4.5 | 11.1 | 32.9 | 24.4 | 38.7 | 50.4 | <5 |
NA: not available. Values above or below reference ranges are marked in bold.
Identification of a mutation in the IRF4 gene
Whole-exome sequencing of DNA samples from P1, his parents, and P2 identified a germline heterozygous missense IRF4 gene variant (Chr6: 401753; hg19 build 137; NM_002460.3, exon 7, c.1075 T>C, p.F359L) located in the protein’s IAD (Fig. 1 E and Fig. S1 B). The mutation was de novo for P1 and inherited for P2 and P3. IRF4’s IAD binds cofactors, such as PU.1 (Brass et al., 1999; Remesh et al., 2015). The variant (confirmed by Sanger sequencing) had a combined annotation-dependent depletion score of 23; this value is well above the mutation significance cutoff reported for the IRF4 gene (Guérin et al., 2018). An earlier analysis of the population genetics indicated that the IRF4 gene has evolved with purifying selection (Guérin et al., 2018). The F359L missense variant’s potential association with the disease was supported by its absence in our in-house database and several open-access human genetic variation databases, including the Exome Aggregation Consortium, the Exome Sequencing Project, and the Genome Aggregation Database. However, the PolyPhen and Sorting Intolerant From Tolerant Prediction tools predicted that the IRF4 F359L missense variant was benign (with scores of 0.174 and 1, respectively; Fig. S1 B). Protein alignments of IRF4 orthologs indicated the evolutionary conservation of the IRF4 IAD as a whole (including the F359 position), with the exception of the zebrafish Danio rerio protein ortholog (Fig. 1 F). Alignment of IADs of the human IRF family members IRF3-9 showed that F359 is conserved in all the assessed IADs (Meraro et al., 1999 and Fig. 1 G). IRF4’s documented role in immune cells, the possible structural/functional importance of amino acid F359, the close segregation of the mutation with the disease in the probands’ family, and the absence of other variants segregating with the disease in genes reported to be associated with PIDs (Bousfiha et al., 2020; Tangye et al., 2020) indicated that the IRF4 F359L variant likely contributed to PAD phenotype.
F359L preferentially alters IRF4 activity at ISRE and AICE promoters
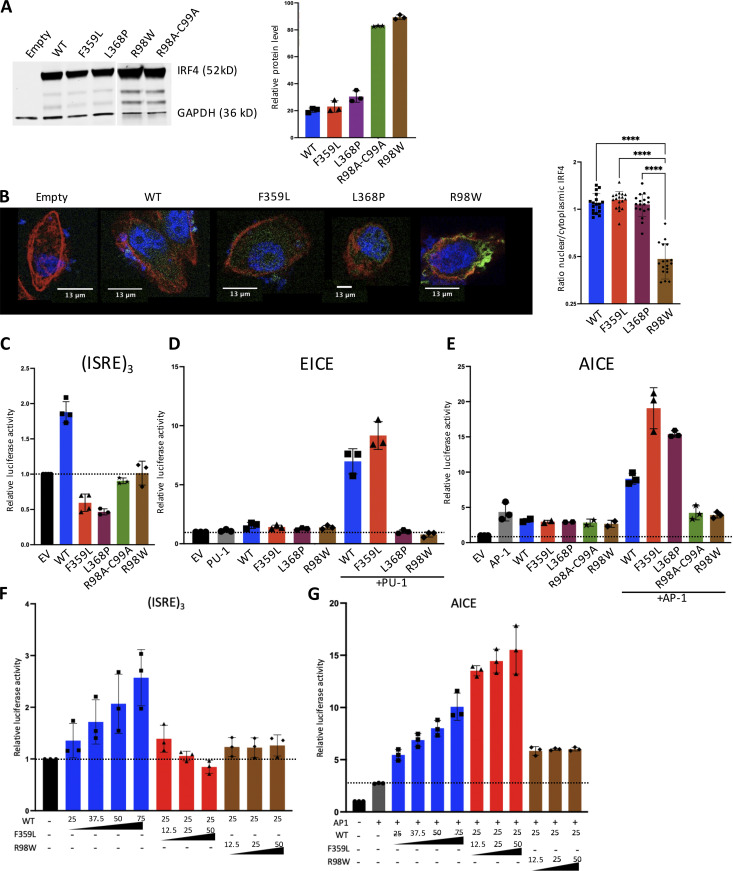
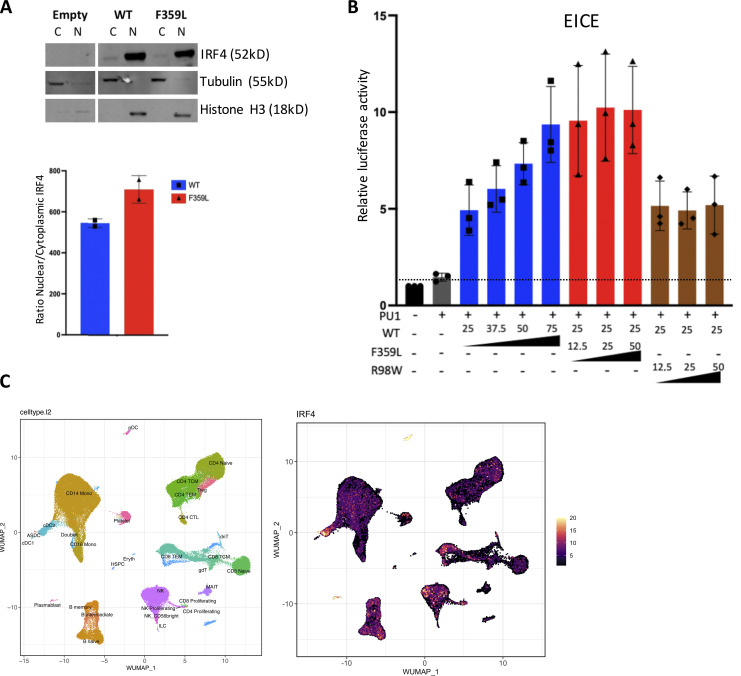
Next, we investigated if the IRF4 F359L variant affects total IRF4 mRNA and protein levels. Sanger sequencing of cDNAs from both T cell blast and patient-derived EBV-immortalized lymphoblastoid B cells (B-EBV cells) suggested that the WT and mutant alleles are likely expressed at similar levels (Fig. 1 H and Fig. S1 C). We found that total IRF4 mRNA expression in T cell blasts was comparable with controls (Fig. 1 I). A Western blot analysis of total cell lysates indicated that IRF4 protein was similarly abundant in patient and control T cell blasts as well as in patient-derived and healthy-donor-derived B-EBV cells (Fig. 1, J–M). To assess the F359L variant’s effect on IRF4 protein function, we transiently expressed either WT IRF4 or IRF4 F359L in HEK293T cells. The previously described IRF4 mutants IRF4 R98A-C99A and R98W (located in the DBD and associated with loss of DNA binding [Brass et al., 1999; Guérin et al., 2018]) and IRF4 L368P (an IRF4 mutant located in the IAD and associated with the loss of ability to interact with PU.1 [Meraro et al., 1999]) were included as controls. Immunoblots of total cell extracts with an anti-IRF4 antibody showed that the IRF4 F359L protein was as abundant as IRF4 WT and IRF4 L368P and less abundant than the DNA-binding-deficient mutants IRF4 R98A-C99A and R98W (Fig. 2 A and Guérin et al., 2018). The IRF4 F359L and IRF4 WT proteins were similar with regard to their subcellular localization in the cytoplasm and the nucleus (Fig. 2 B and Fig. S2 A). The transcriptional function of IRF4 is mediated by ISREs (to which IRF4 binds as a homodimer), EICEs (to which IRF4 binds through an interaction with PU.1), and AICEs (which require a cooperative interaction between IRF4 and the BATF-JUNB heterodimer). To assess IRF4 F359L’s ability to induce transcription by binding to ISRE, EICE, and AICE motifs, we performed the corresponding luciferase reporter assays. Unlike the IRF4 WT protein, all the analyzed mutant IRF4 proteins (F359L, L368P, R98A-C99A, and R98W) failed to activate the (ISRE)3 promoter (Fig. 2 C). It is noteworthy that the amount of luciferase activity was lower for IRF4 proteins F359L and L368P than for IRF4 R98A-C99A and R98W and even the empty vector control. In contrast, IRF4 WT and F359L had similar levels of transcriptional activity via the EICE promoter in the presence of PU.1, whereas IRF4 L368P, R98A-C99A, and R98W failed to activate the EICE promoter (Fig. 2 D). Robust transcriptional activity of IRF4 WT, F359L, and L368P was also observed with the AICE promoter in the presence of AP1 (BATF-JUND heterodimer), with greater activity for IRF4 F359L than for IRF4 WT (Fig. 2 E). These results indicate that IRF4 F359L selectively fails to activate the ISRE promoter. We therefore looked for haploinsufficiency or a dominant-negative effect of the IRF4 F359L protein on the IRF4 WT protein. We found that in the presence of increasing amounts of the IRF4 F359L protein, the IRF4 WT protein failed to efficiently induce transcription via the ISRE promoter (Fig. 2 F). In contrast, increasing amount of IRF4 R98W did not interfere with the IRF4 WT protein’s activity with the ISRE promoter (Fig. 2 F). Taken as a whole, these observations suggest that the IRF4 F359L protein had a dominant-negative effect on the ISRE-motif-containing promoter. It is also noteworthy that the presence of IRF4 F359L protein increased the activity of IRF4 WT protein on the AICE-motif-containing promoter (and to a lesser magnitude) the EICE-motif-containing promoter (Fig. 2 G and Fig. S2 B). These results highlighted a selective, trans-dominant negative effect of IRF4 F359L on the ISRE promoter associated with a gentle gain-of-function on the AICE promoter.
Figure 2.
Functional consequences of the F359L mutation. (A) Total cell lysates of HEK293T cells transfected with the various IRF4 variants were analyzed in Western blots. GAPDH was used as an internal control. IRF4 protein expression was quantified. Data represent mean ± SD of n = 3 independent experiments. (B) Immunofluorescent staining of IRF4 (green) in HELA cells transfected with the various IRF4 mutants. The nucleus was stained with DAPI (blue) and actin filaments in the cytoplasm were stained with phalloidin (red); quantification data represent mean ± SD of 18 cells per condition analyzed within n = 2 independent experiments. A one-way analysis of variance was used to determine whether or not differences were statistically significant (**** P < 0.0001). (C–E) Luciferase activity of HEK293T cells co-transfected with an (ISRE)3 (C), EICE (D), or AICE (E) reporter plasmid plus 75 ng of empty plasmid, or with plasmids encoding the IRF4 variants. (F and G) Luciferase activity of HEK293T cells co-transfected with an (ISRE)3 (F) or AICE (G) reporter plasmid in the presence of 25 ng of IRF4 WT expressing plasmid plus the indicated amount of plasmid encoding the respective IRF4 variants. The quantity of plasmid was normalized to 75 ng by the addition of empty plasmid. For AICE and EICE assays, plasmids encoding BATF and JUN or PU-1 cofactors (25 ng) were added, respectively, under the indicated conditions. Results are shown as the fold-induction in activity, relative to cells transfected with empty plasmid. (C–G) The dotted line indicates the mean level of activity for transfected cells with the empty plasmid. Data represent mean ± SD of n = 3 independent experiments for all conditions depicted in D, F, and G and for IRF4 mutant L368P, R98A-C99A, and R98W in C, mean ± SD n = 4 independent experiments for IRF4 WT, IRF4 F359L, and the empty vector (EV) control are shown in C. (E) Data represent mean ± SD of n = 3 independent experiments for empty vector and all + AP-1 conditions and mean ± SD of n = 2 independent experiments for conditions without AP-1.
Figure S2.
Molecular characterization of IRF4 F359L’s function. (A) Western blot analysis of IRF4’s abundance in nuclear and cytoplasmic cell compartments. HEK293T cells were transfected with an empty plasmid or with plasmids expressing IRF4 WT or IRF4 F359L. Nuclear and cytoplasmic protein extracts were analyzed. Tubulin and histone H3 were respectively used as control cytoplasmic and nuclear proteins. Quantification realized with Image Studio software represents the mean ± SD of n = 2 independent experiments. The Mann–Whitney–Wilcoxon test was used to determine whether or not differences were statistically significant (P < 0.05 was considered statistically significant). (B) Luciferase activity of HEK293T cells co-transfected with an EICE reporter plasmid plus the indicated amount of plasmids encoding the various IRF4 variants. The quantity of plasmid was normalized to 75 ng by addition of empty vector. A plasmid encoding the PU-1 cofactor was added (25 ng) in the indicated conditions. The dotted line indicates the mean level of activity for transfected cells with the empty plasmid. Data represent mean ± SD of n = 3 independent experiments for all conditions depicted. (C) IRF4 expression in the different annotated PBMCs’ populations analyzed by single-cell RNA-Seq. Data extracted from Li et al. (2012).
The immune phenotype of patients indicates impaired plasmablast differentiation and abnormal T cell phenotype
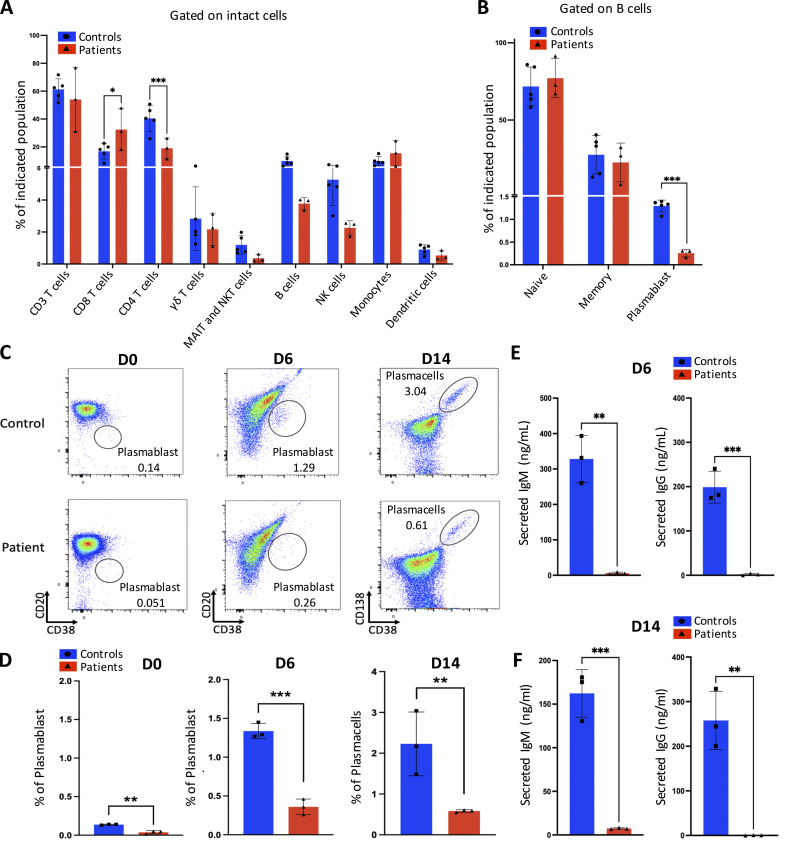
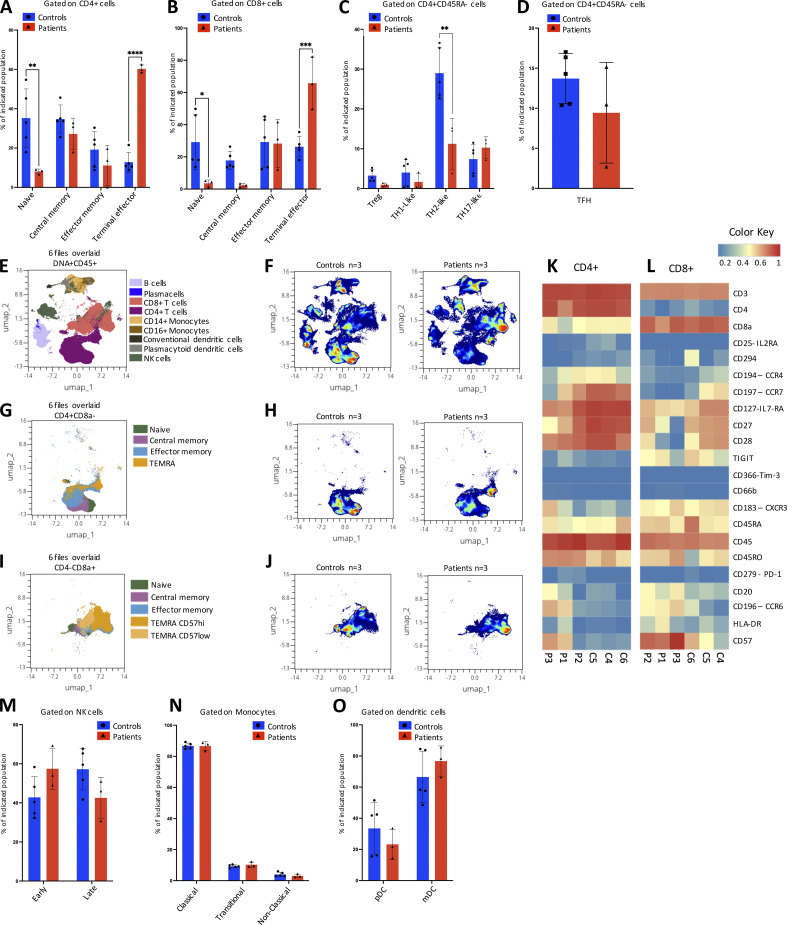
Although all three patients with the IRF4 F359L mutation developed hypogammaglobulinemia early in life, they all displayed normal B cell counts and a normal proportion of switched memory B cells in adulthood (Table 1). A high proportion of CD21low B cells was observed in P2, P3, and at the last evaluation of P1. The absolute T cell counts were low for P1 and P2, whereas high or normal for P3. Similarly, CD4 T cell counts were low for both P1 and P2, whereas variable for P3. In contrast, the CD8 T cell counts were normal or slightly above normal. All three patients had markedly low proportions of naïve CD4 and naïve CD8 T cells and elevated proportions of CD4 and CD8 memory T cells (and especially the memory effector subtype [CD45RA−CCR7−] and/or terminal effector subtype [CD45RA+CCR7−]). Natural killer (NK) cell counts were reported for P1 or normal for P2 and P3. To assess the differential expression of IRF4 transcripts, we analyzed 3′ single-cell RNA sequencing (RNA-Seq) data of the recently published multimodal atlas of human peripheral blood mononuclear cells (PBMCs; Hao et al., 2021). Expression of IRF4 mRNA was observed in all T cell subsets, B cells, NK cells, monocytes, and dendritic cells, with high levels of expression in plasmablasts and plasmacytoid dendritic cells (Fig. S2 C). Subsequent time-of-flight mass cytometry (CyTOF) analysis of PBMCs from all three patients confirmed the normal proportions of naïve and memory B cell populations but revealed a low proportion of plasmablasts in all three individuals—suggesting a defect in plasmablast differentiation (Fig. 3, A and B). To investigate this, we isolated B cells from patients and controls and assessed their ability to differentiate into plasmablasts in vitro after activation. Plasmablast generation (Fig. 3, C and D) and Ig secretion (Fig. 3, E and F) were lost or greatly reduced with the patient’s B cells compared with healthy controls, suggesting an intrinsic defect in differentiation into plasmablasts and plasma cells. For all three patients, the CyTOF analysis also confirmed the very low naïve CD4 and CD8 T cell counts and the markedly elevated counts of CD4 and CD8 terminal effector T cells with high expression of CD57 and CCR6 and low expression of CD127, CD28, CD27, and CCR4; these results further highlighted their terminal differentiation phenotype (Fig. 4, A–L). Inhibitory receptors (like TIGIT, CD279 [PD-1], and CD366 [TIM-3]) were expressed to a similar extent in patient and control samples (Fig. 4, K–L). Similar proportions of CD4 TH17-like, Th1-like, T regulatory, and circulating TFH cells (CXCR5+ cells) were observed in patient and control samples (Fig. 4, C and D). However, a lower proportion of CD4 TH2-like cells was observed in the patient samples (Fig. 4 C). The patients had normal NK, monocyte, dendritic cell, and plasmacytoid dendritic cell proportions (Fig. 4, M–O).
Figure 3.
In-depth immunophenotyping of the patients’ PBMCs and analysis of B cell differentiation. (A and B) Shows results of an automated MaxparPathsetter analysis of CyTOF acquisitions for n = 5 control samples (blue) and n = 3 patient samples (red). Indicated lymphocyte compartments after gating on intact cells (A) and B cell populations after gating on B cells (B) are represented as bar graphs. One-way analyses of variance were used to determine whether or not differences were statistically significant (*** P< 0.001 and * P< 0.05). (C and D) Flow cytometry analysis of (from left to right) the plasmablast subset (CD20−CD38+) or plasma cell subset (CD138+CD38+) at day (D) 0, day 6, or day 14 after activation of enriched B cells from n = 3 controls and n = 3 patients. Representative plots for control and patient samples are shown (C) and bar graph representing the mean ± SD of the analyzed n = 3 control and n = 3 patient samples (D). (E and F) Analysis of secreted IgM (left) or IgG (right) in culture supernatant at D6 (E) or D14 (F); the mean ± SD of the analyzed n = 3 control and n = 3 patient samples are depicted. (D–F) One-way analyses of variance were used to determine whether or not differences were statistically significant (*** P < 0.001 and ** P < 0.01).
Figure 4.
In-depth immunophenotyping of CD4 T cells, NK cells, monocytes, and dendritic cells. (A–C) Results of an automated MaxparPathsetter analysis of CyTOF acquisitions for control samples (blue) and patient samples (red). Percentages of indicated CD4+ T cell subsets (A and C) and CD8+ T cell subsets (B) are represented after gating on intact cells (A and B) or additional gating on CD4+CD45RA− cells (C). (D) TFH cells identified by manual gating as a proportion of CD4+CD45RA− T cells for control samples (blue) and patient samples (red). (A–D) Data represent mean ± SD of n = 5 control samples and n = 3 patient samples. One-way analyses of variance were used to determine whether or not differences were statistically significant (**** P < 0.0001, *** P < 0.001, ** P < 0.01, and * P < 0.05). (E) A uniform manifold approximation and projection (UMAP) obtained by concatenation of n = 3 control samples and n = 3 patient samples using the OMIQ Data Science Platform. The main lymphocyte populations (identified by manual gating) are shown. (F) Density plots of lymphocyte distributions in n = 3 controls (left) and n = 3 patients (right). (G and H) The CD4+ T cell subpopulation represented as a UMAP projection (G) and a density plot (H). (I and J) CD8+ cells subpopulation represented as a UMAP projection (I) and a density plot (J). (K and L) Heatmaps for the expression of surface markers of differentiation on CD4+ T cells (K) and CD8+ T cells (L). Red indicates high expression, yellow indicates intermediate expression, and blue indicates low expression. (M–O) Bar graphs of NK cells (M), monocytes (N), and dendritic cells (O) subpopulations defined by automated MaxparPathsetter analysis. (M–O) Data represent mean ± SD of n = 5 control samples (blue) and n= 3 patient samples (red).
IRF4 F359L impacts the T cell phenotype
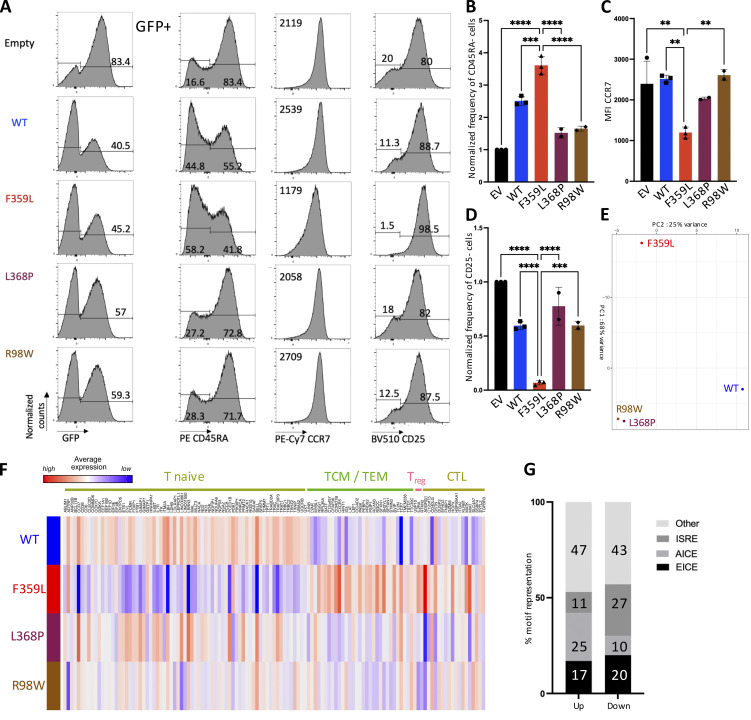
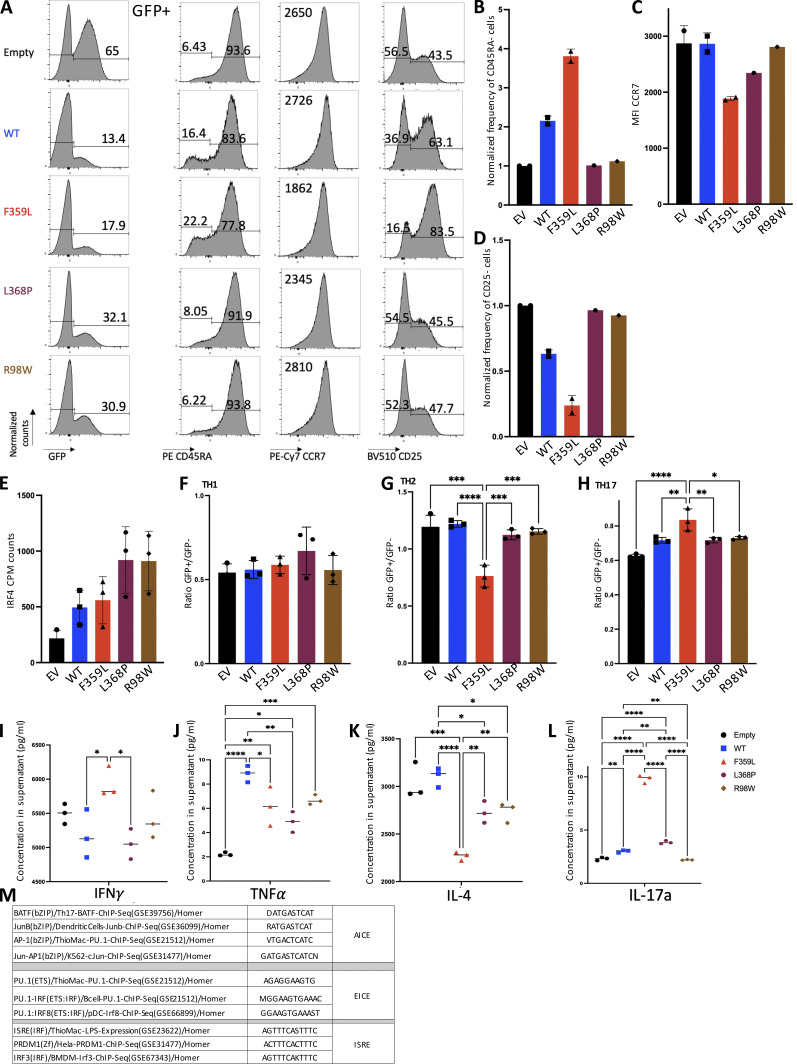
The PBMC immunophenotyping had consistently shown that the patients had low naïve CD4 and CD8 T cell counts and elevated memory CD4 and CD8 T cell counts. We therefore investigated phenotypic changes on naïve CD4 T cells when lentiviral-induced expression of IRF4 F359L was triggered. After flow cytometry sorting, CD4 (CD4+CD45RA+CCR7+) T cells were activated with anti-CD3/-CD28-coupled beads. 3 d later, the cells were infected with lentivirus constructs for GFP alone (the empty vector) or for GFP with IRF4 proteins (the F359L, L368P, and R98W mutants and the WT). An analysis of GFP expression 6 d later indicated that all lentiviral constructs had produced a robust infection (Fig. 5 A). In contrast to cells expressing IRF4 L369P, R98W, or only GFP, cells expressing IRF4 WT and F359L had a lower proportion of CD45RA-negative cells. However, the proportion of CD45RA-negative cells was consistently higher in IRF4-F359L–expressing cells than in IRF4-WT–expressing cells (Fig. 5, A and B). CCR7 expression was slightly lower in IRF4-F359L–expressing cells than in all the other cells (Fig. 5, A and C). In contrast, IRF4-F359L–expressing CD4 T cells included a slightly higher proportion of CD25-positive cells, relative to all other conditions (Fig. 5, A and D). Similar results were obtained for CD8 T cells (Fig. S3, A–D). Taken as a whole, these results indicated that phenotypic changes were induced by the ectopic expression of IRF4 F359L protein in activated naïve CD4 and CD8 T cells.
Figure 5.
Phenotypic changes in naïve CD4 T cells in the presence of IRF4 F359L. (A) Flow cytometry analysis of (from left to right) the GFP signal, and gated on GFP positive cells, CCR7, CD25, or CD45RA expression CD4+ T cells 72 h after transduction with lentiviral vectors expressing (from top to bottom) empty vector, IRF4-WT, IRF4-F359L, IRF4-L368P, or IRF4-R98W. Before activation and transduction, naïve CD4 T cells were sorted (based on CCR7+ and CD45RA+ expression) to obtain a homogenous cell population at the start of the culture. The mean fluorescence intensity (MFI) for CCR7 expression is shown. Representative data for n = 2 independent experiments (IRF4-L368P and IRF4-R98W) or n = 3 independent experiments (Empty, IRF4-WT, IRF4-F359L) are shown. (B–D) Bar graph representing the frequency of CD45RA-negative cells (B), CCR7 MFI (C), and the frequency of CD25-negative cells (B and D are normalized on empty vector [EV] expressing cells). One-way analyses of variance were used to determine whether or not differences were statistically significant (**** P < 0.0001, *** P < 0.001, and ** P < 0.01). (E and F) RNA-Seq analysis of transduced CD4+ T sorted from n = 3 independent healthy controls. Representation of the sample distribution in a PCA (E). (F) Heatmap showing z-score of expressed genes comparing transcripts from IRF4-WT, IRF4-F359L, IRF4-L368P, or IRF4-R98W expressing cells normalized on empty vector expressing CD4+ T cells. Shown genes are top differentially expressed genes for naïve, T cell memory (TCM), T effector memory (TEM), T regulatory (Treg), and cytotoxic lymphocytes (CTL) CD4 subsets identified in Hao et al. (2021). Transcript names are shown along the top axis and associated CD4 subtypes are indicated. (G) Proportion of IRF4-binding sequence motifs from ChIP-Seq peaks extracted from activated CD4+ T cells analysis (GSM2810038) associated with differentially expressed genes. Binding sequences were determined in a HOMER known de novo motif analysis. Motifs are summarized in Fig. S3 M.
Figure S3.
Phenotypic changes in T cells in the presence of IRF4 F359L. (A) Flow cytometry analysis of (from left to right) the GFP signal, and gated on GFP positive cells, CCR7, CD25, and CD45RA expression in CD8+ T cells 72 h after transduction with lentiviral vectors expressing (from top to bottom) empty vector, IRF4-WT, IRF4-F359L, IRF4-L368P, or IRF4-R98W. Before activation and transduction, naïve CD8 T cells were sorted (based on CCR7+ and CD45RA+ expression) to obtain a homogenous cell population at the start of the culture. The mean fluorescence intensity (MFI) for CCR7 expression is shown. Data are representative of n = 2 independent experiments (empty, IRF4-WT, and IRF4-F359L) or n = 1 experiment (IRF4-L368P and IRF4-R98W). (B–D) A bar graph showing (from left to right) the frequency of CD45RA-negative cells (B), CCR7 MFI (C), and the frequency of CD25-negative cells (D); B and D are normalized on empty vector (EV) expressing cells. Data represent mean ± SD of n = 2 independent experiments for empty vector, IRF4-WT, and IRF4-F359L conditions and n = 1 experiment for IRF4-L368P and IRF4-R98W conditions. (E) IRF4 transcript count per million mapped reads (CPM) in CD4+ T cells infected with lentivirus constructs for IRF4 proteins (the F359L, L368P, and R98W mutants and the WT) in comparison with the empty vector. Data represent mean ± SD of n = 3 independent experiments. (F–L) Naive CD4+ T cells polarization toward Th1, Th2, or Th17 subtypes after transduction with lentivirus constructs for IRF4 proteins (the F359L, L368P, and R98W mutants and the WT) or the empty vector. The polarization was performed on three independent healthy donors’ cells. Bar graph representing the percentage of Th1 (F), Th2 (G), or Th17 (H) cells identified in the GFP+ population normalized on the proportion found in the GFP− population. Data represent mean ± SD of n = 3 independent experiments for all conditions depicted. (F–H) One-way analyses of variance were used to determine whether or not differences were statistically significant (**** P < 0.0001, *** P < 0.001, ** P < 0.01, and * P < 0.05). (I and J) Analysis of secreted IFNγ (I) or TNFa (J) in supernatant of the Th1 culture. (K) Analysis of secreted IL-4 in supernatant of the Th2 culture. (L) Analysis of secreted IL-17a in supernatant of the Th17 culture. (I–L) Data represent mean ± SD of n = 3 independent experiments for all conditions depicted. One-way analyses of variance were used to determine whether or not differences were statistically significant (**** P < 0.0001, *** P < 0.001, ** P < 0.01, and * P < 0.05). (M) Motifs taken into account for the annotation of ISRE, EICE, and AICE sites in the HOMER analysis.
To investigate the IRF4 F359L mutation’s functional impact on the regulation of transcription in activated naïve CD4 T cells, we performed RNA-Seq to analyze mRNA expression in cells infected with lentivirus constructs 6 d after infection. Elevated IRF4 transcript count per million mapped reads was observed in cells infected with lentivirus constructs for IRF4 proteins (the F359L, L368P, and R98W mutants and the WT) in comparison with the empty vector (Fig. S3 E). The RNA-Seq profiles were normalized to the empty vector condition to allow integration of the different experiments performed with different healthy donor samples. A principal component analysis (PCA) of transcriptomic data indicated clear differences between cells expressing the IRF4 F359L mutant protein and cells expressing either IRF4 L368P and R98W mutants or the IRF4 WT (Fig. 5 E). Increased expression of transcripts for marker genes (top differentially expressed genes in the CD4 T cell subset of the multimodal atlas of human PBMCs [Hao et al., 2021]) for CD4 T central memory, effector memory, regulatory, and CD4+ T cells with cytotoxic activity was observed in the F359L expressing cells compared with cells expressing IRF4 WT, or either IRF4 L368P or R98W mutants. In contrast, lower expression of transcripts for CD4 T naïve marker genes was observed in cells expressing the IRF4 F359L mutant protein compared with cells expressing IRF4 WT (Fig. 5 F). To identify the DNA binding motifs associated with the deregulation in gene expression, we intersected publicly available anti-IRF4 chromatin immunoprecipitation sequencing (ChIP-Seq) data from human CD4 T cells (GSM2810038) and our RNA-Seq data. In a motif analysis using HOMER software (http://homer.ucsd.edu/homer/), we analyzed AICE, EICE, and ISRE motifs (Fig. S3 M) in the ChIP-Seq peaks for genes showing increased and decreased expression in the presence of IRF4 F359L mutant protein compared with cells expressing IRF4 WT. Genes that were more strongly expressed in IRF4 F359L cells had a greater number of AICE-containing motifs (Fig. 5 G). In contrast, genes that were less expressed in the IRF4 F359L cells had a greater number of ISRE-containing motifs (Fig. 5 G). Taken as a whole, these results indicate that binding of IRF4 F359L to AICE sites in CD4 T cells was correlated with higher expression of a set of genes, some of which higher expressed in CD4 T central memory, effector memory, regulatory, and CD4+ T cells with cytotoxic activity. Since the CyTOF analysis of patients' PBMCs indicated a lower proportion of CD4 TH2-like cells, we investigated polarization of activated naïve CD4 T cells toward CD4 TH2-like when lentiviral-induced expression of IRF4 F359L was triggered. Evaluation of chemokine receptors and cytokine secretion of CD4 T cells expressing IRF4 F359L, IRF4 WT, IRF4 L368P, or IRF4 R98W protein cultured under polarizing conditions indicated no impact of IRF4 F359L on polarization to TH1-like cells, whereas IRF4 F359L impaired polarization to TH2-like and increased polarization to TH17-like cells when compared with IRF4 WT and either IRF4 L368P or R98W mutant condition was observed (Fig. S3, F–L).
IRF4 F359L impairs plasmablast/plasma cell generation
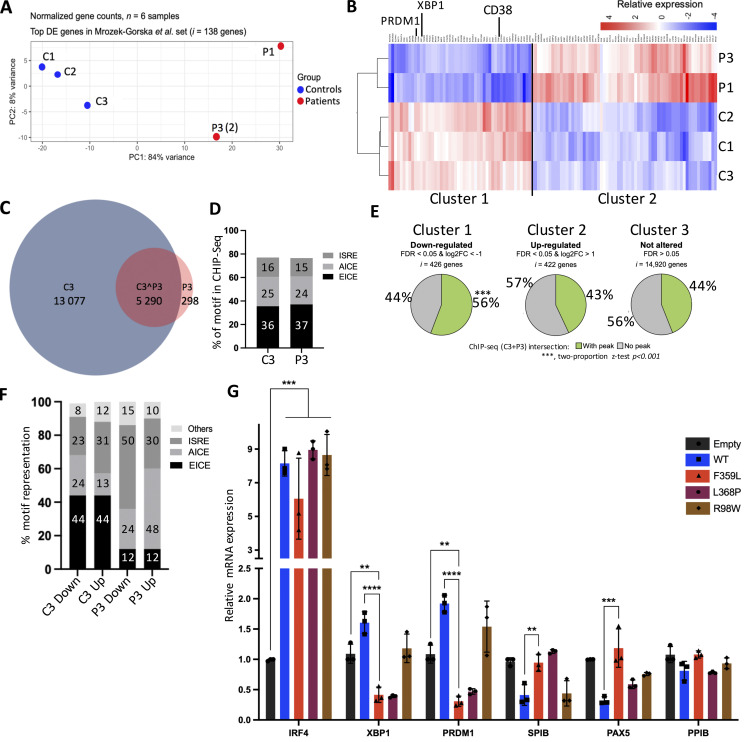
It has been reported that EBV infection immortalizes human B lymphocytes; the latter proliferate and have much the same transcriptomic profile as early plasma cells (Mrozek-Gorska et al., 2019). To investigate the IRF4 F359L mutation’s functional impact on the regulation of transcription, we used RNA-Seq to analyze mRNA expression in the patient- and donor-derived B-EBV cell lines. For P3, two B-EBV cell lines derived from two independent blood samples were analyzed. A PCA of transcriptomic data for genes marking various steps in the differentiation of mature B cells into early plasma cells (the “immune gene” transcriptome list for the top 138 differentially expressed genes in Mrozek-Gorska et al., 2019) highlighted clear differences between patient cells and control cells (Fig. 6 A). Since the RNA-Seq profiles of both of P3’s independent B-EBV cell lines clustered together within the PCA, we used the mean of the two for all subsequent analyses. Hierarchical clustering of the changes in expression in healthy donor B-EBV cells vs. patient cells identified three clusters: cluster I contained 426 genes expressed more strongly in control samples than in patient samples; cluster II contained 422 genes expressed more strongly in patient samples than in control samples (Fig. 6 B); and cluster III contained 14,920 genes expressed to a similar extent in control and patient samples. Interestingly, genes associated with the terminal differentiation of activated B cells into plasma cells (including PRDM1, XBP1, and CD38) were found in cluster I—suggesting that plasma blast/plasma cell gene expression program was altered in patients B-EBV lines (Fig. 6 B).
Figure 6.
IRF4 F359L–expressing B-EBV cell lines present a plasmablast/plasma cell differentiation defect. (A and B) RNA-Seq analysis of patient and control B-EBV cell lines; in total n = 6 samples, n = 3 controls, and n = 3 patient samples. Patient samples were obtained from different blood samples (n = 1 for P1 and n = 2 for P3). Representation of the sample distribution in a PCA of 138 genes (the top differentially expressed [DE] genes in Mrozek-Gorska et al., 2019) (A) Heatmap of 115 genes expressed differentially when comparing control (n = 3) B-EBV cells with patient (n = 2) B-EBV cells. (B) Mean expression of (n = 2) B-EBV cell lines is shown for P3. Transcript names are shown along the top axis, and key genes in plasma cell differentiation are highlighted. Clusters 1 and 2, respectively, encompass genes that are downregulated or upregulated in patient cells. (C and D) ChIP-Seq analysis of patient (n = 1) and control (n = 1) B-EBV cell lines. Venn diagrams represent the distribution of ChIP-Seq peaks present in both the control and patient samples (C). Proportion of peaks in control and patient samples detected for the different IRF4 binding sequences determined in a HOMER known de novo motif analysis (D). (E) Correlation between RNA gene expression and IRF4 binding (the respective ChIP-Seq peaks of patient and control samples were compared). Two proportion z statistical analyses were performed (*** P < 0.001). (F) Proportion of IRF4-binding sequence motifs (determined in a HOMER known de novo motif analysis) from ChIP-Seq peaks of differentially expressed genes. (G) qPCR analysis of selected indicated transcripts. Data represent mean ± SD of n = 3 different control B-EBV cells transduced with lentiviral vectors expressing IRF4 WT or various IRF4 mutant proteins. One-way analyses of variance were used to determine whether or not differences were statistically significant (**** P < 0.0001, *** P < 0.001, and ** P < 0.01).
It has been reported that a shift in IRF4 binding from AICE sites to ISRE sites is associated with the IRF4 protein concentration and the pattern of B lymphocyte differentiation (Cocco et al., 2019 Preprint; Ochiai et al., 2013). To analyze IRF4’s binding to regulatory elements, we performed ChIP-Seq on both patient and healthy donor (control) B-EBV cell lines. Overall, the number of IRF4-bound regions was lower in the patient sample than in the control sample (Fig. 6 C). In a motif analysis using HOMER software, however, the IRF4-bound regions in patient and control cells had similar distributions of AICE, EICE, and ISRE motifs (Fig. 6 D and Fig. S3 M). It is noteworthy that the number of IRF4-bound sites within proximal promoter was lower in the patient cells than in control cells (Fig. S4 A). Next, we intersected the anti-IRF4 ChIP-Seq data and the RNA-Seq data. Gene transcripts that were similarly expressed or upregulated in the patient B-EBV cells (relative to control cells) displayed similar frequencies of ChIP-Seq peaks (44 and 43%, respectively; Fig. 6 E), whereas gene transcripts that were downregulated in patient cells (relative to controls) had a greater frequency of ChIP-Seq peaks (56%); these findings suggest that IRF4 F359L’s binding to chromatin impaired gene expression (Fig. 6 E). To identify the DNA binding motifs associated with the impairment in gene expression, we analyzed the motifs in the ChIP-Seq peaks for genes in cluster I and cluster II. Genes that were more strongly expressed in control cells had a greater number of ISRE-containing motifs (Fig. 6 F). In contrast, genes that were less expressed in the patient cells had a greater number of ISRE-containing motifs (Fig. 6 F). Of note, genes that were more strongly expressed in patient cells had a greater number of AICE-containing motifs (Fig. 6 F). Taken as a whole, these results indicate that binding of IRF4 F359L to ISRE sites was correlated with low expression of a set of genes, some of which were involved in plasma cell differentiation. Furthermore, the results of the motif analysis were in line with the low observed level of transcription on the ISRE-driven promoter and the high observed level of transcription on the AICE-driven promoter induced by IRF4 F359L in HEK293T cells (Fig. 2, C and F).
Figure S4.
ChIP-Seq and RIME analyses. (A) The genome-wide distribution of IRF4 ChIP-Seq peaks in B-EBV cells. The locations of ChIP-Seq peaks associated with differentially expressed genes in control and patient samples (based on the RNA-Seq analysis) were annotated. (B) Peptide coverage of the IRF4 protein in RIME. Peptides recognized are highlighted in gray. High proportions of identified peptides are indicated by blue lines. (C) Evaluation of shRNA mediated knockdown of ETV6 in HEK293T cells by quantitative RT-PCR. Data represent mean ± SD of n = 3 independent experiments. One-way analyses of variance were used to determine whether or not differences were statistically significant (**** P < 0.0001 and ** P < 0.01). (D) Western blotting. (E) Quantification of D realized with Image Studio software. (D and E) Data represent mean ± SD of n = 3 independent experiments. One-way analyses of variance were used to determine whether or not differences were statistically significant (*** P < 0.001 and * P < 0.05).
We next looked at whether or not expression of IRF4 F359L protein interfered with the transcription of XBP1 and PRDM1. Both genes are involved in plasma cell differentiation and were found in cluster I. Using a lentiviral vector, control B-EBV cells were transduced so that they expressed either GFP alone (the empty vector) or GFP with IRF4 proteins (WT, F359L, L368P, or R98W). The transduced cells were sorted (based on GFP expression) and the transcripts of IRF4, XBP1, PRDM1, PAX5, SPIB, and PPIB were quantified. Relative to all other conditions, cells transduced with IRF4 F359L expressed lower levels of XBP1 and PRDM1 transcripts. In contrast, cells expressing IRF4 F359L and cells expressing GFP only had similar levels of SPIB, PAX5, and PPIB transcripts (Fig. 6 G). It is noteworthy that the transcription levels of SPIB and PAX5 were lower in cells expressing IRF4 WT ectopically than in cells expressing GFP only or IRF4 F359L. Taken as a whole, these results demonstrate that IRF4 F359L interferes with the expression of PRDM1 and XBP1 and further support the hypothesis whereby IRF4 F359L expression impairs the differentiation of activated B cells into plasma cells.
F359L alters IRF4 binding to transcription cofactors and chromatin-associated proteins
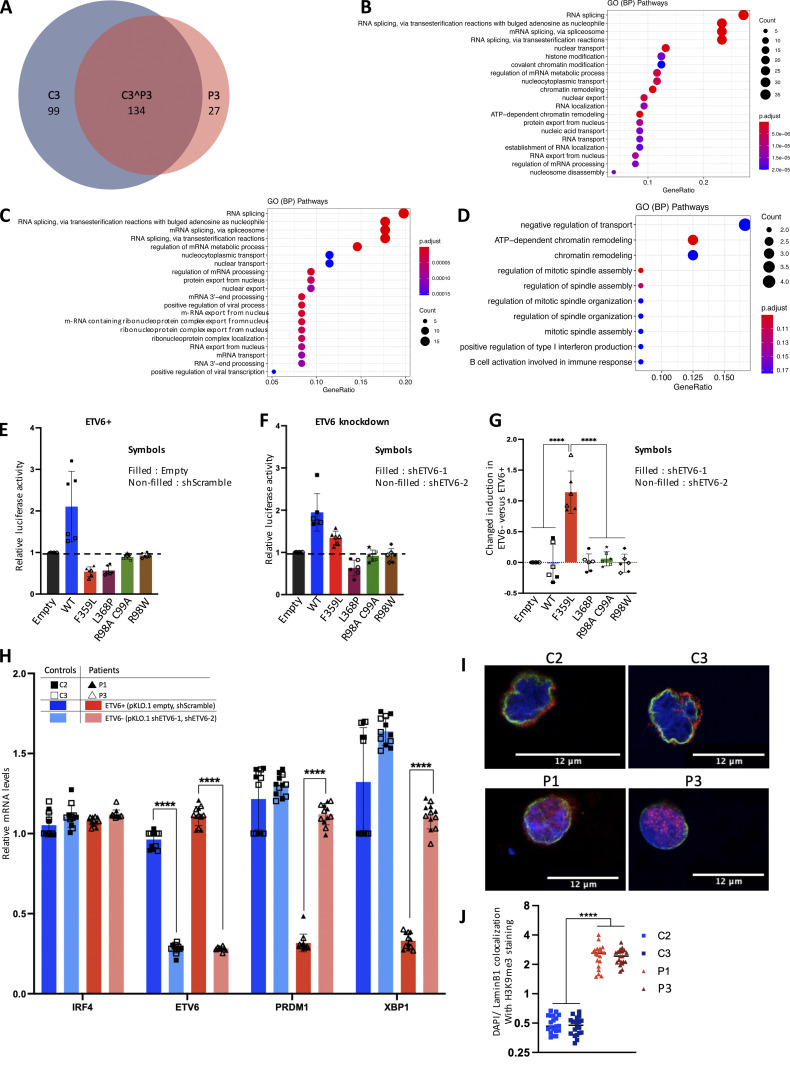
To investigate the IRF4 F359L mutation’s impact on binding to transcription cofactors and chromatin-associated proteins, we performed rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME; Mohammed et al., 2013) experiments on both patient and healthy donor (control) B-EBV cell lines. The immunoprecipitated protein complexes showed a good level of coverage (55–60%) of the IRF4 bait protein (Fig. S4 C). The RIME analysis reproducibly detected 233 IRF4-associated proteins in the control sample and 161 IRF4-associated proteins in the patient sample. A total of 134 proteins were common to both datasets (Fig. 7 A and Table S1), including IRF4-interacting proteins annotated in the BioGRID interaction database (e.g., TOP1, FKBP4, and ARM1) or the Reactome Functional Interaction network (SMARCC1, SMARCC2, and SMARCA4). The gene ontology terms for these components were related to RNA processing (including transport, localization, and splicing) and chromatin remodeling (Fig. 7 B). Our analysis also showed that 99 proteins were reproducibly detected in the control sample but not in the patient (Fig. 7 A and Table S1); the gene ontology terms of these proteins were related to RNA splicing and nuclear pore organization (Fig. 7 C). Conversely, 27 components were detected in the patient sample but not in the control sample (Fig. 7 A and Table S1); the related gene ontology terms were positive regulation of type I interferon production, and ATP-dependent chromatin remodeling (Fig. 7 D). It is noteworthy that ETV6 (one of the proteins detected solely in the patient sample, also known as TEL) has been linked to transcriptional regulation (repression) at ISRE sites (Kuwata et al., 2002). No differences in mRNA expression were observed for 95 (96%) of the 99 IRF4-immunoprecipitated, chromatin-associated proteins detected solely in control samples, and 25 (93%) of the 27 proteins detected solely in patient samples. This finding suggests strongly that the distinct detection patterns were related to bona fide modifications in interaction with the IRF4 F359L mutant, rather than differences in protein expression. Thus, the RIME analysis revealed that the chromatin-bound IRF4 F359L protein interacts with a partially different set of cell components.
Figure 7.
Disturbance of the IRF4 protein’s interactions with chromatin, functional impact of ETV6, and localization of trimethylated (K9) histone H3 in patient B-EBV cells. (A–D) The results of an IRF4 RIME analysis in patient B-EBV cells (P3, n = 1) and control (C3, n = 1) B-EBV cells. The Venn diagram shows the distribution of reproducibly identified proteins in n = 2 independent experiments (A). (B–D) A gene ontology biological process pathway analysis for protein interactions with IRF4 on chromatin in both control and patient B-EBV cell samples (B), in the control sample only (C), or in the patient sample only (D). (E–G) Luciferase activity; data represent mean ± SD of control (shScramble n = 3, empty vector n = 3; E) or ETV6 knockdown (shETV6-1 n = 3, shETV6-2 n = 3; F) infected HEK293T cells co-transfected with an (ISRE)3 reporter plasmid plus 75 ng of empty plasmid, or with plasmids encoding the IRF4 variants. (G) Changed induction of luciferase activity due to ETV6 knockdown for the different IRF4 variants. The results present the relative luciferase activity determined in ETV6 deficient cells (presented in F)—relative luciferase activity determined in ETV6 proficient cells (presented in E). One-way analyses of variance were used to determine whether or not differences were statistically significant (**** P < 0.0001). (H) qPCR analysis of selected indicated transcripts from n = 2 control and n = 2 patient B-EBV cell lines transduced with lentiviral vectors expressing for control conditions (shScramble, empty vector) or ETV6 knockdown (shETV6-1, shETV6-2). Three independent experiments were performed. Data represent mean ± SD of n = 12 samples for ETV6+ control cells, n = 12 samples for ETV6+ patient cells, n = 12 samples for ETV6− control cells, and n = 12 samples for ETV6− patient cells. One-way analyses of variance were used to determine whether or not differences were statistically significant (**** P < 0.0001). (I) Representative images of immunofluorescent staining of H3K9me3 (red) in n = 2 control and n = 2 patient B-EBV cell lines. The nucleus was stained with DAPI (blue) and the inner nuclear membrane was stained for lamin B1 (green). (J) Shown is quantification of I (using Fiji software) in 40 cells per condition from a total of n = 2 independent experiments; line indicates median. One-way analyses of variance were used to determine whether or not differences were statistically significant (**** P < 0.0001).
To assess whether ETV6 was responsible for IRF4 F359L’s inability to activate the (ISRE)3 promoter, we performed luciferase reporter assays in short hairpin RNA (shRNA) mediated ETV6 knockdown HEK293T cells (Fig. 7, E and F). Efficient knockdown of ETV6 (by two different shRNA) was confirmed on RNA and protein levels (Fig. S4, C–E). The IRF4 F359L protein was active on the (ISRE)3 promoter in ETV6 knockdown cells albeit to a lower level compared with IRF4 WT protein (Fig. 7 F). It is noteworthy that ETV6 knockdown affected only the capability of IRF4 F359L to activate the (ISRE)3 promoter but had no significant impact on other analyzed mutant proteins (WT, L368P, R98A-C99A, and R98W; Fig. 7 G). To investigate whether ETV6 is responsible for the suppressed gene expression of PRDM1 and XBP1 in patients’ B-EBV cell lines, we knocked down ETV6 in these cells. In patients’s ETV6 knockdown B-EBV cell lines, we observed an increased gene expression of PRDM1 and XBP1 compared with patients’s B-EBV cell lines without ETV6 knockdown (Fig. 7 H). Taken as a whole, these results demonstrate that the transcriptional repressor ETV6 interferes with the activity of IRF4 F359L on ISRE sites.
IRF4 was found to bind to the positive coactivator 4 (PC4; encoded by SUB1) in the control sample but not in the patient sample. It is noteworthy that a PC4 protein complex containing IRF4 has been described in activated murine B cells (Ochiai et al., 2020). Given that PC4 has been linked functionally to the positioning of heterochromatin (Ochiai et al., 2020), we next analyzed the subcellular localization of histone H3 K9 trimethylation (H3K9me3, a heterochromatin marker) in control and patient B-lymphoblastoid cell lines. In control cells, the H3K9me3 signal colocalized with lamin B1 at the boundary of the nucleus (Fig. 7, H–I). In contrast, the H3K9me3 signal in patient cells was spread throughout the nucleus. Overall, these results indicate that the interaction between heterochromatin and the nuclear membrane is impaired in the presence of IRF4 F359L.
Discussion
Here, we reported on three patients from a multigenerational family carrying a private heterozygous missense variant of IRF4 (c.1075 T>C, p.F359L). The inheritance pattern and the results of our functional analysis suggest that the IRF4 F359L variant causes a novel PAD that manifests itself as hypogammaglobulinemia with T cell abnormalities and hair and skin pigmentation anomalies. The disease segregated in an autosomal dominant manner and appeared to have complete penetrance; it therefore differs from a previously reported IRF4 deficiency, i.e., a combined immunodeficiency characterized by agammaglobulinemia, eosinophilia, normal lymphocyte counts, low memory T and B cell counts, and elevated GM-CSF–induced macrophage polarization (Bravo García-Morato et al., 2018) and IRF4 haploinsufficiency with incomplete, age-dependent penetrance (caused by a heterozygous loss-of-function mutation affecting IRF4’s DBD and the clinical manifestations of which are due to Tw exposure [Guérin et al., 2018]).
The IRF4 F359L mutation is located within the IAD—a well-defined, evolutionarily conserved structural domain involved in homo- and heterodimer protein–protein interactions (Remesh et al., 2015; Sundararaj et al., 2021). Functional IRF4 forms in the context of high protein abundance and ISRE sites homodimer and binds to EICE and AICE sites as a heterodimer (Cook et al., 2020). Our ChIP-Seq data indicated that the IRF4 WT protein and the mutant IRF4 F359L protein bound to ISRE, EICE, and AICE motifs to similar extents. While the results of our experiments with various luciferase reporter constructs showed that IRF4 F359L did not negatively impact AICE- and EICE-dependent promoter activities, mutant IRF4 F359L was not able to activate ISRE-driven promoters and even exerted a dominant-negative effect on IRF4 WT. These findings indicate that structural changes in IRF4 F359L do not negatively interfere with binding to ISRE, EICE, and AICE motifs nor with the interaction of PU1 and AP1, but do interfere with the IRF4-driven transcriptional activity through ISRE. We even observed a gentle gain-of-function of IRF4 F359L on AICE promoter in the presence of AP1 (BATF-JUND heterodimer).
Apart from hypogammaglobulinemia diagnosed early in life, a very low plasmablast/plasma cell count in the blood was the only consistent B lineage defect observed in the three patients. The observed normalization of IgA titers with age might be related to the accumulation of long-lived plasma cells over time (Manz et al., 2005). Defective in vitro differentiation of purified B cells as well as low RNA expression of PRDM1, XBP1, and CD38 detected by RNA-Seq in B-EBV cell lines indicated a likely intrinsic defect in plasmablast/plasma cell differentiation. This is consistent with the known association between IRF4-driven expression of PRDM1 and a conserved, non-coding region containing ISRE binding sites within the gene (Sciammas et al., 2006). We also showed that the ectopic expression of IRF4 F359L in B-EBV cell lines derived from healthy individuals repressed PRDM1 and XBP1 gene expression. Furthermore, the combined analysis of RNA expression and IRF4 ChIP-Seq data indicated the presence of a greater number of ISRE sites in genes that were expressed less strongly in patient cells than in control cells. These data can be interpreted as an ISRE-promoted dominant-negative effect of IRF4 F359L’s transcriptional activity.
Many of the IRF4 partner proteins identified in the RIME analysis are known to be components of (i) a complex with PC4 (encoded by Sub1 [Ochiai et al., 2020]) and (ii) switch/sucrose non-fermentable remodeling complexes (Centore et al., 2020); this observation indicates that functional IRF4 not only acts as a transcriptional activator but also is an important factor in chromatin remodeling and chromosome organization. The IRF4 F359L protein might hinder the interaction with components involved in nuclear pore organization (these components were identified predominantly in the control RIME sample) and probably disturbs chromatin remodeling, chromosome organization, and chromatin accessibility. This hypothesis is supported by the altered localization of the H3K9me3 signal with the nuclear membrane in patient cells. The transcriptional regulators ETV6 and ARID1B were detected solely in patient cells. ETV6 reportedly interacted with IRF8 (Humblin et al., 2017) at ISRE sites in particular (Kuwata et al., 2002). Our (ISRE)3 promoter luciferase reporter assays in ETV6 knockdown cells demonstrated that IRF4 F359L’s binding to ETV6 is at least partly responsible for the dominant-negative impact on ISRE-motif-mediated transcriptional regulation. This notion is further supported by the increased gene expression of PRDM1 and XBP1 in patients’ B-EBV cell lines due to ETV6 knockdown.
Studies of murine models have suggested that IRF4 senses TCR signaling strength (Krishnamoorthy et al., 2017) or acts as a “rheostat” by translating TCR affinity into the appropriate transcriptional programs (Man et al., 2013). These studies also found that IRF4 promotes CD8 T cell exhaustion (Man et al., 2017). Accordingly, our observation with human cells indicated that IRF4 F359L promotes terminal differentiation into CD4 and CD8 T cell subsets. Firstly, the IRF4 F359L patients had abnormal T cells, with low naïve CD4 and CD8 T cell counts and elevated proportions of terminal effector CD4 and CD8 T cells. Secondly, the ectopic expression of mutant IRF4 F359L in naïve CD4 and CD8 T cells was associated with transcriptomic changes and lower proportions of CD45RA-negative cells, relative to expression in non-transduced cells and control cells. The combined analysis of RNA expression and public available IRF4 ChIP-Seq data indicated the presence of a greater number of AICE sites in genes that were expressed more strongly in IRF4 F359L expressing cells than in control cells. These data can be interpreted as an AICE-promoted gain-of-function effect of IRF4 F359L’s transcriptional activity reminding of the gentle gain-of-function of IRF4 F359L on AICE promoter in the presence of AP1 (BATF-JUND heterodimer). A gain-of-function of IRF4 F359L on AICE sites could possibly be explained by an enhanced IRF4 F359L–JUND (or other JUN family member) protein interaction, since JUNB protein was one of the 27 components detected in the patient samples but not in the control samples in our RIME analysis. Taken together, our phenotyping and functional data suggest that the dysfunctional IRF4 F359L protein accelerates the differentiation of naïve T cells into terminal effector memory T cells and impairs the differentiation of B cells into plasma cells. However, the low overall naïve B and naïve T cell counts in patients might indicate that IRF4 F359L not only impacts the T and B cell differentiation processes but also interferes with early B and T cell development and/or the homeostasis of naïve B and naïve T cells. Splenomegaly and lymphadenopathy (due to the expansion of both T and B lymphocytes) have been reported in 10–15-wk-old Irf4-deficient mice, which suggests that IRF4 has a functional role in the homeostasis of both B and T lymphocytes (Mittrücker et al., 1997). It has also been suggested that IRF4 has a functional role in the homeostasis of mature B cells (their positioning in lymphoid microenvironments, to be precise; Simonetti et al., 2013).
The results of genome-wide association studies have suggested a role for IRF4 in pigmentation (Praetorius et al., 2013) and in hair graying (Adhikari et al., 2016). The premature hair graying observed in our three patients (none of whom carried the rs12203592 SNP) strongly suggests that the process was mediated by IRF4 F359L dysfunction.
Several PIDs are known to be caused by heterozygous missense variants in transcription factors (Bousfiha et al., 2020; Tangye et al., 2020) via gain-of-function, negative dominance, haploinsufficiency, or heterodimeric interference (Yamashita et al., 2021). Our data fit with a novel disease-causing role for IRF4 F359L. This mutant enhances AICE-motif-mediated transcription and interferes with ISRE-motif-mediated transcriptional regulation in a dominant negative manner. Our evidence of an interaction between IRF4 F359L and ETV6 in the context of ISRE sites suggests that this neomorphic mutation causes an autosomal dominant disease with a novel disease mechanism: protein function is impaired by a change in binding partners and/or functional interference between protein complexes, rather than by the loss- or gain-of-function of individual proteins.
Material and methods
Blood sample collection from patients and healthy donors, and study approval
Peripheral blood samples were collected from the patients after the provision of written, informed consent. Genetic studies and data collection procedures were approved by the local institutional review board (Comité de Protection des Personnes Ile de France II, Paris, France; reference: 2015-01-05; 2015-01-05 MS2) and the French Advisory Committee on Data Processing in Medical Research (Comité Consultatif sur le Traitement de l’Information en matière de Recherche dans le domaine de la Santé, Paris, France; reference: 15.297bis).
Whole-exome sequencing
Whole-exome sequencing and analysis were performed as described previously (Bouafia et al., 2019). Exome capture was performed using the SureSelect Human All Exon Kit (Agilent Technologies). Agilent SureSelect Human All Exon (58 Mb, V6) libraries were prepared from 3 μg of genomic DNA sheared with an ultrasonicator (Covaris), as recommended by the manufacturer. Barcoded exome libraries were pooled and sequenced using a HiSeq 2500 system (Illumina) to generate paired-end reads. After demultiplexing, sequences were mapped against the human genome reference (NCBI build37/hg19 version) with the Burrows-Wheeler Aligner (version 0.7.12; Li and Durbin, 2009). The mean depth of coverage obtained for the two exome libraries exceeded 150×, and more than 97% and more than 96% of the targeted exonic bases were covered by at least 15 and 30 independent sequencing reads, respectively (≥97% at ×15 and ≥96% at ×30). Variants were called with the Genome Analysis Toolkit (GATK), SAMtools, and Picard Tools. SNPs were called with the GATK Unified Genotyper, whereas indels were called with the GATK IndelGenotyper, version 2. All variants with a read coverage of ×2 or less and a Phred-scaled quality of 20 or less were filtered out. All the variants were annotated and filtered using PolyWeb (our in-house annotation software).
Cell culture
Using SepMate PBMC Isolation Tubes (#85450; STEMCELL Technologies) and Ficoll, we isolated PBMCs from cytopheresis rings (from healthy volunteers) or whole-blood samples (from patients). PBMCs and B-EBV cells were cultured in RPMI medium supplemented with 10% fetal bovine serum, whereas HEK293T cells were cultured in DMEM medium supplemented with 10% fetal bovine serum. T cell blasts were generated by stimulation of PBMCs with PMA/ionomycin (#tlrl-pma; Invivogen and #I0634; Sigma-Aldrich) and cultured in Xvivo 15 supplemented with 10% human serum (type AB) (Sigma-Aldrich) and interleukin-2 (300 IU/ml).
Plasmablast differentiation assay
PBMCs isolated from healthy donors’ or patients’ blood were thawed, and B cell enrichment was performed using both Pan B cell isolation and dead cell removal kits (#130-101-638, #130-090-101; Miltenyi). B cells were seeded at 50,000 cells per well in 96-well plate in RPMI medium supplemented with 10% fetal bovine serum containing IL2 (100 UI/ml), IL4 (100 ng/ml), IL21 (100 ng/ml), IL10 (25 ng/ml), CD40L (200 ng/m), and anti IgM antibody (5 µg/ml). Medium was renewed every 3 d. Cell differentiation was analyzed by flow cytometry and Ig secretion by ELISA (#BMS2098, #BMS2091; Thermo Fisher Scientific) on day 6 and day 14.
Plasmids
Full-length constructs carrying mutant alleles were generated from pcDNA 3.1D/V5-His-TOPO IRF4 WT (Guérin et al., 2018) using the GeneArt Site-Directed Mutagenesis System (#A13282; Thermo Fisher Scientific), according to the manufacturer’s instructions. We used the lentiviral pWPI backbone provided by Addgene. All the constructs (WT, F359L, L368P, and R98W IRF4) were generated by GenScript. The SFR BioSciences Gerland-Lyon Sud (Lyon, France) vector facility produced the lentiviral supernatant, and a multiplicity of infection of 30 was used for both CD4, CD8, and B-EBV cells.
For ETV6 knockdown experiments, shRNA expressing lentiviral constructs (Sigma-Aldrich, TRCN0000003853 [shETV6-1] and TRCN0000003856 [shETV6-2]) and corresponding controls (SHC016-1EA; Sigma-Aldrich [shScramble] and 10878; Addgene [Empty vector]) were used.
Transfection
HEK293T and HELA cells were transfected transiently with the various constructs by using Lipofectamine 2000 (Thermo Fisher Scientific) in accordance with the manufacturer’s instructions.
Cell lysis and Western blotting
Total protein extracts were prepared by incubating cells on ice for 45 min with lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.5% Triton X-100, and 2 mM EDTA) supplemented with protease and phosphatase inhibitors (#87786; Thermo Fisher Scientific). In a two-step procedure, the cytoplasmic and nuclear contents were separated using Thermo Fisher Scientific NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific) according to the manufacturer’s instructions. Equal amounts of protein (according to a Bradford protein assay [Thermo Fisher Scientific, Micro BCA Protein Assay Kit]) were resolved by SDS-PAGE in a NuPAGE 10%, Bis-Tris gel (Invitrogen) and transferred to a low-fluorescence polyvinylidene difluoride membrane. Membranes were probed with nonconjugated antibody: an anti-IRF4 (#4948; Cell Signaling) antibody was used at a dilution of 1:1,000, ETV6 (#PA5-81865; Thermo Fisher Scientific), and antibodies against GAPDH (sc-32233; Santa Cruz), tubulin (T5168; Sigma-Aldrich), and histone H3 (#9715; Cell Signaling) were used at the same dilution as loading controls. Antibodies bound to the membrane were detected by incubation with the appropriate infrared-dye-conjugated secondary antibody (Li-Cor, 926-68071, 926-32211 and 926-32210) in a Licor Odyssey CLx system (Li-Cor). Images were analyzed and quantified with Image Studio Lite software.
Immunofluorescent staining in B-EBV cells
B-EBV cells were harvested and fixed by incubation with 4% paraformaldehyde for 15 min and permeabilized in a 10 min incubation with 1× PBS 5% BSA/0.1% Triton X-100. Cells were washed with filtered PBS, blocked by a 40 min incubation with PBS 5% BSA, stained for IRF4 (#4948; Cell Signaling), H3K9me3 (#710816; Thermo Fisher Scientific), and Lamin B1 (#sc-377000; Santa Cruz) for 1 h, washed, and incubated with a secondary antibody (anti-rabbit FITC Jackson, #50784, donkey anti-rabbit #A21206; Invitrogen, and goat anti-mouse #A11003; Invitrogen). DAPI (#910-3018; Chemometech) and phalloidin (#8940; Cell Signaling) were added for 40 min. After several washes, the cells were placed on precoated coverslips with poly-D-lysine (#A3890401; Gibco). Fluorescence was detected with a confocal Leica SP8 microscope. Images were analyzed with Fiji software.
Luciferase reporter assays
The (ISRE)3 reporter plasmid (containing three repeats of the ISRE sequence separated by spacers), the AICE reporter plasmid, and the EICE reporter plasmid have been described elsewhere (Doody et al., 2007; Guérin et al., 2018). HEK293T cells were transiently transfected with the (ISRE)3 reporter plasmid (100 ng/well in a 96-well plate), the pRL-SV40 vector (# E2231, 40 ng/well; Promega), and an IRF4 WT or mutant pcDNA 3.1D/V5-His-TOPO plasmid (#K4900-01; Invitrogen, 75 ng or the amount indicated, together with an empty plasmid to give 75 ng) and Lipofectamine 2000 (#11668019; Thermo Fisher Scientific), according to the manufacturer’s instructions. The same protocol was used for the AICE and EICE assays, except that cofactor expression plasmids (respectively BATF and JUN or PU-1) were added (25 ng/well). Cells were analyzed 24 h after transfection using the Dual-Luciferase 1,000 assay system kit (#E1980; Promega) and according to the manufacturer’s instructions. Signal intensity was determined with an EnVision multimode plate reader (Perkin Elmer). Experiments were performed in triplicate, and reporter activity was expressed as the fold-induction relative to cells transfected with the empty vector. Negative dominance or haploinsufficiency was assessed in cells transfected with a constant amount of WT plasmid (25 ng/well), various amounts of mutant plasmid (from 12.5 to 50 ng/well, together with empty plasmid to give a total of 75 ng), (ISRE)3 reporter plasmid (100 ng/well for a 96-well plate), and pRL-SV40 vector (40 ng/well). The same protocol was used for the AICE and EICE luciferase reporter assays, except that cofactor expression plasmids (25 ng/well each) were added.
RNA-seq and quantitative PCR (qPCR) assays
Total RNA was prepared from the B-EBV cells of individuals with a heterozygous IRF4 mutation (two patients) from healthy, homozygous WT individuals (n = 4). RNA was prepared (including a DNase treatment step) from 500,000 cells by using the RNeasy Plus Kit (Qiagen), according to the manufacturer’s instructions.
For RNA-Seq, RNA quality was assessed by capillary electrophoresis with high-sensitivity RNA reagents and a Fragment Analyzer (Agilent Technologies). The RNA concentration was measured spectrophometrically with an Xpose system (Trinean) and by using capillary electrophoresis (Fragment Analyzer). RNA-Seq libraries were prepared from an initial total of 200 ng RNA using the Universal Plus mRNA-Seq kit (Nugen), as recommended by the manufacturer (libraries can be prepared from 10 ng to 1 µg of starting total RNA). The oriented cDNAs produced from the poly-A+ fraction were sequenced on a NovaSeq6000 system (Illumina: paired-end reads 100 bases + 100 bases). Around 50 million filtered paired-end reads were produced for each library. Reads were aligned with the hg38 human reference genome using HISAT2 (Kim et al., 2019), and those that mapped uniquely to GENCODE-annotated genes were summarized using featureCounts (Liao et al., 2014). The raw gene count matrix was imported into the R environment (https://www.R-project.org/) for further processing and analysis. Genes with low read counts (less than ∼10 reads in more than three samples) were filtered out, leaving a set of ∼18,000 genes to be tested for differential expression in healthy homozygous WT individuals (control) vs. individuals with heterozygous IRF4 mutations (patients). Read counts were normalized and differential expression was analyzed by applying three independent, complementary statistical methods: DESeq2 (Love et al., 2014), edgeR (Robinson et al., 2010), and Limma-voom (Law et al., 2014). We applied the optimal procedure, which consisted in simultaneously filtering the analysis results with a statistical significance threshold (Benjamini-Hochberg adjusted P values <5%) and a fold-change threshold (|fold change| >2). Z-score was calculated as follows: ((gene expression value in sample of interest) − (mean expression across all samples))/(standard deviation) was used as a scaling method for visualization in heatmaps.
For quantitative real-time PCR, a High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific) was used to generate cDNA. Quantitative real-time PCR was performed with TaqMan Universal PCR Master Mix (Thermo Fisher Scientific), specific primers (all from Thermo Fisher Scientific) for IRF4 (Hs00180031_m1), XBP1 (Hs00231936_m1), PRDM1 (Hs00153357_m1), SPIB (Hs00162150_m1), PPIB (Hs00168719_m1), PAX5 (Hs00277134_m1), ETV6 (Hs00231101_m1), and (as a control) endogenous human GAPDH (Hs02786624_g1) or HBS1L (Hs04188641_g1). The data were analyzed using the ΔΔCt method, with normalization against GAPDH.
CyTOF phenotyping of PBMCs
A combination of the Maxpar Direct Immune Profiling Assay (#201325; Fluidigm) and the Maxpar Direct T cell Expansion Panel 2 (#201406; Fluidigm) was used for the high-dimensional immune profiling of PBMCs. The antibody markers analyzed are summarized in Table S2. For each sample, cells were thawed, washed once, and checked for viability (>80%). After a 10 min incubation with FC Blocker (Biolegend, TruStain FCX) in MaxPar staining buffer, cells were directly transferred into the antibody-containing tube. Cells were then incubated for 10 min in 1.6% formaldehyde solution, washed once, transferred into Intercalator ID solution, and incubated overnight at 4°C. Immediately before acquisition, the cells were washed, resuspended in Maxpar Cell Acquisition Solution (1 million cells per ml), and mixed with 10% vol/vol EQ Beads. An average of 500,000 events were acquired per sample on a Helios mass cytometer. The acquisition data were analyzed with CyTOF software (Fluidigm, version 6.7.1014), enabling MaxparPathsetter automated single-cell analysis at the Cytometry Facility in Pitié-Salpetriere Hospital (Paris, France) or by manual gating with the OMIQ analysis platform. Data were analyzed with either standardized MaxparPathsetter automated single-cell analysis, and populations were defined following markers’ expression given in Bagwell et al. (2020) or by unsupervised clustering with manual identification; in this case, population was defined following markers’ expression given in Geanon et al. (2020 Preprint).
Analysis of phenotypic changes in naïve T cells expressing IRF4 F359L
CD4+ and CD8+ naïve T cells were isolated from healthy donor PBMCs and then labeled with anti-CD4 (#345768; BD), anti-CD8 (#345773; BD), anti-CD45RA (#304138; Biolegend), and anti-CCR7 (#130-099-363; Miltenyi) antibodies. Naïve T cells (defined as CD45RA+ CCR7+ CD4+ cells) were isolated (>98% purity) with a FACS AriaII cell sorter (BD Biosciences). Purified naive CD4+ or CD8+ T cells were activated with a Dynabeads T cell activation kit (anti-CD3/CD28; #11131D; Gibco) and cultured in a medium containing interleukin-2 (300 IU/ml). After 24 h, the cells were transduced using adjuvant (Lentiboost, Sirion Biotech, and PGE2 Cayman, #14750) and lentiviral vectors (pWPI backbone) expressing WT or mutant IRF4. For T cell polarization 1 d after transduction, cytokines were added to the culture as indicated: TH1 polarization IL12 (50 ng/ml), TH2 polarization IL4 (100 U/ml), TH17 polarization TGFβ (2.5 ng/ml), IL1β (20 ng/ml), IL6 (50 ng/ml), IL21 (50 ng/ml), IL23 (100 ng/ml), and PGE2 (50 ng/ml). Using flow cytometry, we evaluated the transduction efficiency, viability, and differentiation 6 d after transduction, on the basis of GFP expression, a fixable viability dye (Thermo Fisher Scientific), and surface staining with anti-CD25 (#563352; BD), anti-CCR7 (#557648; BD), and anti-CD45RA (#304112; Biolegend), or anti-CCR6 (#565925; BD), CCR7 (#557648; BD), CXCR3 (#353706; Biolegend), and CCR4 (#557863; BD) antibodies, respectively. Cytokine secretion was evaluated using Human Th1/Th2/Th17 CBA Kit (#560484; BD) following the protocol given by the provider.
ChIP-Seq
B-EBV cells (10 × 106) from P3 and a healthy control C1 were fixed with freshly prepared 1% formaldehyde (#F-8775; Sigma-Aldrich) for 15 min, quenched with 0.125 M glycine (#G-7403; Sigma-Aldrich) for 5 min, and washed twice with 1× PBS containing 0.5% Igepal CA-630 (#I-8896; Sigma-Aldrich) and (in the second wash only) 1 mM phenylmethanesulfonyl fluoride. Cell pellets were snap-frozen and subjected to ChIP-Seq. Chromatin extraction, immunoprecipitation with an anti-IRF4 antibody (#4948; Cell Signaling), library preparation, next-generation sequencing, and a model-based analysis of the ChIP-Seq data (Zhang et al., 2008) were performed by Active Motif. Motifs were discovered using HOMER software (http://homer.ucsd.edu/homer/) with its default parameters.
RIME
B-EBV cells (50 × 106) from P3 and a healthy control C1 were fixed with 1% formaldehyde for 15 min, quenched with 0.125 M glycine, and snap-frozen. RIME experiments (Mohammed et al., 2016) in B-EBV cells were performed by Active Motif using an anti-IRF4 antibody (#4948; Cell Signaling) or an isotype-matched IgG. The RIME analysis was performed in replicates. Proteins present in both experimental replicates with a spectral count of ≥5 (upon filtering the experimental reaction data against the negative control IgG reaction data) were included into the final protein list.
Induction of IRF4 F359L expression in control B-EBV cells
Healthy-donor-derived B-EBV cells were transduced using an adjuvant (Lentiboost, Sirion Biotech) and lentiviral vectors (pWPI backbone) expressing WT or mutant IRF4 proteins. 6 d later, transduced cells were isolated (>98% purity) by cell sorting with an AriaII cell sorter (BD Biosciences) on the basis of their GFP expression. Total RNA was extracted.
ETV6 knockdown
HEK 293T expressing WT or mutant IRF4 proteins or healthy-donor-/patient-derived B-EBV cells were transduced using an adjuvant (Sirion Biotech, Lentiboost) and lentiviral vectors (pLKO.1 backbone). 3 d later, transduced cells were selected by the addition of puromycin. 4 d later, knockdown cells were selected and used for further analysis.
Statistics
Differences between populations were probed using a one-way analysis of variance. All analyses were performed with GraphPad Prism software (version 9.2.0, https://www.graphpad.com). The threshold for statistical significance was set to P < 0.05.
Online supplemental material
The supplementary information shows clinical features and genetic analysis (Fig. S1), further molecular characterization of IRF4 F359L’s function (Fig. S2), additional exploration of phenotypic changes in T cells in the presence of IRF4 F359L (Fig. S3), and further information concerning ChIP-Seq and RIME analyses (Fig. S4). Table S1 lists the proteins found in RIME experiments, and Table S2 lists the antibodies used in CyTOF experiments.
Supplementary Material
shows proteins found in RIME experiments.
shows the antibodies used in CyTOF experiments.
Acknowledgments
We thank the patients and their families for contributing to the study. We thank Dr. Marion Espeli, Dr. Vivien Béziat, and Dr. Pierre Milpied for helpful discussions. We thank the Necker Imagine Centre de Resources Biologiques for generating EBV-transformed lymphoblastoid cell lines; the clinical research team at the Imagine Institute for their support; the Etablissement Français du Sang for blood supply from healthy donors; SIRION Biotech GmbH for providing LentiBOOST; Gisèle Froment, Didier Nègre, and Caroline Costa from the lentivector production facility at SFR BioSciences Gerland - Lyon Sud (UMS3444/CNRS/US8/INSERM, École normale supérieure de Lyon, Université Claude Bernard Lyon 1) for viral vector production; and Dr. Gina Doody (University of Leeds, Leeds, UK) for providing us with the EICE reporter plasmid.
The work was funded by the French state (via the Agence Nationale de la Recherche’s “Investissments d’avenir” program [ANR-10-IAHU-01], and grants ANR-19-CE17-0012-01, ANR-19-CE17-0012-02, ANR-19-CE17-0012-03, and ANR-19-CE17-0012-04 [ANR-AID]), the Ligue Contre le Cancer–Comité de Paris, and INSERM. S. Kracker is a Centre National de la Recherche Scientifique staff researcher.
Author contributions: R. Thouenon and S. Kracker designed and analyzed experiments. R. Thouenon, L. Chentout, N. Moreno-Corona, and L. Poggi performed and analyzed experiments. All authors contributed to data analysis. S. Kracker supervised research. E. Puig Lombardi performed computational analysis. R. Thouenon, L. Chentout, and Y. Schmitt performed whole-exome sequencing and RNA-Seq experiments. R. Thouenon and B. Hoareau performed CyTOF experiments. L. Galicier, J. Fadlallah, and A. Fischer provided patient care and clinical information. C. Lagresle-Peyrou, J.-L. Casanova, and J. Bustamante provided reagents. A. Fischer, A. Durandy, L. Galicier, and J. Fadlallah provided samples from patients and controls. S. Kracker and R. Thouenon wrote the manuscript, and all the co-authors revised, edited, and approved the manuscript.
Data availability
The RNA-Seq data of lentiviral transduced CD4 T cells can be accessed in the Gene Expression Omnibus (GEO) repository under the accession number GSE214888. The ChIP-Seq and RNA-Seq data of B-EBV cells can be accessed in the GEO repository under the accession numbers GSE199684 and GSE199685 (GSE199686 SuperSeries).
References
- Adhikari, K., Fontanil T., Cal S., Mendoza-Revilla J., Fuentes-Guajardo M., Chacón-Duque J.-C., Al-Saadi F., Johansson J.A., Quinto-Sanchez M., Acuña-Alonzo V., et al. 2016. A genome-wide association scan in admixed Latin Americans identifies loci influencing facial and scalp hair features. Nat. Commun. 7:10815. 10.1038/ncomms10815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagwell, C.B., Hunsberger B., Hill B., Herbert D., Bray C., Selvanantham T., Li S., Villasboas J.C., Pavelko K., Strausbauch M., et al. 2020. Multi-site reproducibility of a human immunophenotyping assay in whole blood and peripheral blood mononuclear cells preparations using CyTOF technology coupled with Maxpar Pathsetter, an automated data analysis system. Cytometry B Clin. Cytom. 98:146–160. 10.1002/cyto.b.21858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouafia, A., Lofek S., Bruneau J., Chentout L., Lamrini H., Trinquand A., Deau M.-C., Heurtier L., Meignin V., Picard C., et al. 2019. Loss of ARHGEF1 causes a human primary antibody deficiency. J. Clin. Invest. 129:1047–1060. 10.1172/JCI120572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousfiha, A., Jeddane L., Picard C., Al-Herz W., Ailal F., Chatila T., Cunningham-Rundles C., Etzioni A., Franco J.L., Holland S.M., et al. 2020. Human inborn errors of immunity: 2019 update of the IUIS phenotypical classification. J. Clin. Immunol. 40:66–81. 10.1007/s10875-020-00758-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass, A.L., Kehrli E., Eisenbeis C.F., Storb U., and Singh H.. 1996. Pip, a lymphoid-restricted IRF, contains a regulatory domain that is important for autoinhibition and ternary complex formation with the Ets factor PU.1. Genes Dev. 10:2335–2347. 10.1101/gad.10.18.2335 [DOI] [PubMed] [Google Scholar]
- Brass, A.L., Zhu A.Q., and Singh H.. 1999. Assembly requirements of PU.1-Pip (IRF-4) activator complexes: Inhibiting function in vivo using fused dimers. EMBO J. 18:977–991. 10.1093/emboj/18.4.977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo García-Morato, M., Aracil Santos F.J., Briones A.C., Blázquez Moreno A., Del Pozo Maté Á., Domínguez-Soto Á., Beato Merino M.J., Del Pino Molina L., Torres Canizales J., Marin A.V., et al. 2018. New human combined immunodeficiency caused by interferon regulatory factor 4 (IRF4) deficiency inherited by uniparental isodisomy. J. Allergy Clin. Immunol. 141:1924–1927.e18. 10.1016/j.jaci.2017.12.995 [DOI] [PubMed] [Google Scholar]
- Centore, R.C., Sandoval G.J., Soares L.M.M., Kadoch C., and Chan H.M.. 2020. Mammalian SWI/SNF chromatin remodeling complexes: Emerging mechanisms and therapeutic strategies. Trends Genet. 36:936–950. 10.1016/j.tig.2020.07.011 [DOI] [PubMed] [Google Scholar]
- Cocco, M., Care M.A., Al-Maskari M., Doody G., and Tooze R.. 2019. A dichotomy in association of core transcription factors and gene regulation during the activated B-cell to plasmablast transition (preprint). bioRxiv. (Preprint posted December 23, 2019). 10.1101/2019.12.23.884007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, S.L., Franke M.C., Sievert E.P., and Sciammas R.. 2020. A synchronous IRF4-dependent gene regulatory network in B and helper T cells orchestrating the antibody response. Trends Immunol. 41:614–628. 10.1016/j.it.2020.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva, N.S., Simonetti G., Heise N., and Klein U.. 2012. The diverse roles of IRF4 in late germinal center B-cell differentiation. Immunol. Rev. 247:73–92. 10.1111/j.1600-065X.2012.01113.x [DOI] [PubMed] [Google Scholar]
- Doody, G.M., Stephenson S., McManamy C., and Tooze R.M.. 2007. PRDM1/BLIMP-1 modulates IFN-γ-dependent control of the MHC class I antigen-processing and peptide-loading pathway. J. Immunol. 179:7614–7623. 10.4049/jimmunol.179.11.7614 [DOI] [PubMed] [Google Scholar]
- Durandy, A., Kracker S., and Fischer A.. 2013. Primary antibody deficiencies. Nat. Rev. Immunol. 13:519–533. 10.1038/nri3466 [DOI] [PubMed] [Google Scholar]
- Fusaro, M., Rosain J., Grandin V., Lambert N., Hanein S., Fourrage C., Renaud N., Gil M., Chevalier S., Chahla W.A., et al. 2021. Improving the diagnostic efficiency of primary immunodeficiencies with targeted next-generation sequencing. J. Allergy Clin. Immunol. 147:734–737. 10.1016/j.jaci.2020.05.046 [DOI] [PubMed] [Google Scholar]
- Geanon, D., Lee B., Kelly G., Handler D., Upadhyaya B., Leech J., Herbinet M., Del Valle D., Gnjatic S., Kim-Schulze S., et al. 2020. A streamlined CyTOF workflow to facilitate standardized multi-site immune profiling of COVID-19 patients. medRxiv. (Preprint posted June 29, 2020). 10.1101/2020.06.26.20141341 [DOI] [Google Scholar]
- Guérin, A., Kerner G., Marr N., Markle J.G., Fenollar F., Wong N., Boughorbel S., Avery D.T., Ma C.S., Bougarn S., et al. 2018. IRF4 haploinsufficiency in a family with Whipple’s disease. Elife. 7:e32340. 10.7554/eLife.32340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, J., Kraft P., Nan H., Guo Q., Chen C., Qureshi A., Hankinson S.E., Hu F.B., Duffy D.L., Zhao Z.Z., et al. 2008. A genome-wide association study identifies novel alleles associated with hair color and skin pigmentation. PLoS Genet. 4:e1000074. 10.1371/journal.pgen.1000074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, Y., Hao S., Andersen-Nissen E., Mauck W.M. III, Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M., et al. 2021. Integrated analysis of multimodal single-cell data. Cell. 184:3573–3587.e29. 10.1016/j.cell.2021.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber, M., and Lohoff M.. 2014. IRF4 at the crossroads of effector T-cell fate decision. Eur. J. Immunol. 44:1886–1895. 10.1002/eji.201344279 [DOI] [PubMed] [Google Scholar]
- Humblin, E., Thibaudin M., Chalmin F., Derangère V., Limagne E., Richard C., Flavell R.A., Chevrier S., Ladoire S., Berger H., et al. 2017. IRF8-dependent molecular complexes control the Th9 transcriptional program. Nat. Commun. 8:2085. 10.1038/s41467-017-01070-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D., Paggi J.M., Park C., Bennett C., and Salzberg S.L.. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37:907–915. 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, U., Casola S., Cattoretti G., Shen Q., Lia M., Mo T., Ludwig T., Rajewsky K., and Dalla-Favera R.. 2006. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat. Immunol. 7:773–782. 10.1038/ni1357 [DOI] [PubMed] [Google Scholar]
- Krishnamoorthy, V., Kannanganat S., Maienschein-Cline M., Cook S.L., Chen J., Bahroos N., Sievert E., Corse E., Chong A., and Sciammas R.. 2017. The IRF4 gene regulatory module functions as a read-write integrator to dynamically coordinate T Helper cell fate. Immunity. 47:481–497.e7. 10.1016/j.immuni.2017.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwata, T., Gongora C., Kanno Y., Sakaguchi K., Tamura T., Kanno T., Basrur V., Martinez R., Appella E., Golub T., and Ozato K.. 2002. Gamma interferon triggers interaction between ICSBP (IRF-8) and TEL, recruiting the histone deacetylase HDAC3 to the interferon-responsive element. Mol. Cell. Biol. 22:7439–7448. 10.1128/MCB.22.21.7439-7448.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law, C.W., Chen Y., Shi W., and Smyth G.K.. 2014. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15:R29. 10.1186/gb-2014-15-2-r29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., and Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25:1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, P., Spolski R., Liao W., Wang L., Murphy T.L., Murphy K.M., and Leonard W.J.. 2012. BATF-JUN is critical for IRF4-mediated transcription in T cells. Nature. 490:543–546. 10.1038/nature11530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, Y., Smyth G.K., and Shi W.. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30:923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Love, M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man, K., Gabriel S.S., Liao Y., Gloury R., Preston S., Henstridge D.C., Pellegrini M., Zehn D., Berberich-Siebelt F., Febbraio M.A., et al. 2017. Transcription factor IRF4 promotes CD8+ T cell exhaustion and limits the development of memory-like T cells during chronic infection. Infect. Immun. 47:1129–1141.e5. 10.1016/j.immuni.2017.11.021 [DOI] [PubMed] [Google Scholar]
- Man, K., Miasari M., Shi W., Xin A., Henstridge D.C., Preston S., Pellegrini M., Belz G.T., Smyth G.K., Febbraio M.A., et al. 2013. The transcription factor IRF4 is essential for TCR affinity-mediated metabolic programming and clonal expansion of T cells. Nat. Immunol. 14:1155–1165. 10.1038/ni.2710 [DOI] [PubMed] [Google Scholar]
- Manz, R.A., Hauser A.E., Hiepe F., and Radbruch A.. 2005. Maintenance of serum antibody levels. Annu. Rev. Immunol. 23:367–386. 10.1146/annurev.immunol.23.021704.115723 [DOI] [PubMed] [Google Scholar]
- Meraro, D., Hashmueli S., Koren B., Azriel A., Oumard A., Kirchhoff S., Hauser H., Nagulapalli S., Atchison M.L., and Levi B.Z.. 1999. Protein-protein and DNA-protein interactions affect the activity of lymphoid-specific IFN regulatory factors. J. Immunol. 163:6468–6478 [PubMed] [Google Scholar]
- Mittrücker, H.-W., Matsuyama T., Grossman A., Kündig T.M., Potter J., Shahinian A., Wakeham A., Patterson B., Ohashi P.S., and Mak T.W.. 1997. Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science. 275:540–543. 10.1126/science.275.5299.540 [DOI] [PubMed] [Google Scholar]
- Mohammed, H., D’Santos C., Serandour A.A., Ali H.R., Brown G.D., Atkins A., Rueda O.M., Holmes K.A., Theodorou V., Robinson J.L.L., et al. 2013. Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep. 3:342–349. 10.1016/j.celrep.2013.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed, H., Taylor C., Brown G.D., Papachristou E.K., Carroll J.S., and D’Santos C.S.. 2016. Rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) for analysis of chromatin complexes. Nat. Protoc. 11:316–326. 10.1038/nprot.2016.020 [DOI] [PubMed] [Google Scholar]
- Mrozek-Gorska, P., Buschle A., Pich D., Schwarzmayr T., Fechtner R., Scialdone A., and Hammerschmidt W.. 2019. Epstein-Barr virus reprograms human B lymphocytes immediately in the prelatent phase of infection. Proc. Natl. Acad. Sci. USA. 116:16046–16055. 10.1073/pnas.1901314116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam, S., and Lim J.-S.. 2016. Essential role of interferon regulatory factor 4 (IRF4) in immune cell development. Arch. Pharm. Res. 39:1548–1555. 10.1007/s12272-016-0854-1 [DOI] [PubMed] [Google Scholar]
- Negishi, H., Ohba Y., Yanai H., Takaoka A., Honma K., Yui K., Matsuyama T., Taniguchi T., and Honda K.. 2005. Negative regulation of Toll-like-receptor signaling by IRF-4. Proc. Natl. Acad. Sci. USA. 102:15989–15994. 10.1073/pnas.0508327102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiai, K., Kondo H., Okamura Y., Shima H., Kurokochi Y., Kimura K., Funayama R., Nagashima T., Nakayama K., Yui K., et al. 2018. Zinc finger-IRF composite elements bound by Ikaros/IRF4 complexes function as gene repression in plasma cell. Blood Adv. 2:883–894. 10.1182/bloodadvances.2017010413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiai, K., Maienschein-Cline M., Simonetti G., Chen J., Rosenthal R., Brink R., Chong A.S., Klein U., Dinner A.R., Singh H., and Sciammas R.. 2013. Transcriptional regulation of germinal center B and plasma cell fates by dynamical control of IRF4. Immunity. 38:918–929. 10.1016/j.immuni.2013.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiai, K., Yamaoka M., Swaminathan A., Shima H., Hiura H., Matsumoto M., Kurotaki D., Nakabayashi J., Funayama R., Nakayama K., et al. 2020. Chromatin protein PC4 orchestrates B cell differentiation by collaborating with IKAROS and IRF4. Cell Rep. 33:108517. 10.1016/j.celrep.2020.108517 [DOI] [PubMed] [Google Scholar]
- Praetorius, C., Grill C., Stacey S.N., Metcalf A.M., Gorkin D.U., Robinson K.C., Van Otterloo E., Kim R.S.Q., Bergsteinsdottir K., Ogmundsdottir M.H., et al. 2013. A polymorphism in IRF4 affects human pigmentation through a tyrosinase-dependent MITF/TFAP2A pathway. Cell. 155:1022–1033. 10.1016/j.cell.2013.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remesh, S.G., Santosh V., and Escalante C.R.. 2015. Structural studies of IRF4 reveal a flexible autoinhibitory region and a compact linker domain. J. Biol. Chem. 290:27779–27790. 10.1074/jbc.M115.678789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, M.D., McCarthy D.J., and Smyth G.K.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciammas, R., Shaffer A.L., Schatz J.H., Zhao H., Staudt L.M., and Singh H.. 2006. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 25:225–236. 10.1016/j.immuni.2006.07.009 [DOI] [PubMed] [Google Scholar]
- Simonetti, G., Carette A., Silva K., Wang H., De Silva N.S., Heise N., Siebel C.W., Shlomchik M.J., and Klein U.. 2013. IRF4 controls the positioning of mature B cells in the lymphoid microenvironments by regulating NOTCH2 expression and activity. J. Exp. Med. 210:2887–2902. 10.1084/jem.20131026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundararaj, S., Seneviratne S., Williams S.J., Enders A., and Casarotto M.G.. 2021. Structural determinants of the IRF4/DNA homodimeric complex. Nucleic Acids Res. 49:2255–2265. 10.1093/nar/gkaa1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangye, S.G., Al-Herz W., Bousfiha A., Chatila T., Cunningham-Rundles C., Etzioni A., Franco J.L., Holland S.M., Klein C., Morio T., et al. 2020. Human inborn errors of immunity: 2019 update on the classification from the international union of immunological societies expert committee. J. Clin. Immunol. 40:24–64. 10.1007/s10875-019-00737-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita, M., Kuehn H.S., Okuyama K., Okada S., Inoue Y., Mitsuiki N., Imai K., Takagi M., Kanegane H., Takeuchi M., et al. 2021. A variant in human AIOLOS impairs adaptive immunity by interfering with IKAROS. Nat. Immunol. 22:893–903. 10.1038/s41590-021-00951-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., and Liu X.S.. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol. 9:R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows proteins found in RIME experiments.
shows the antibodies used in CyTOF experiments.
Data Availability Statement
The RNA-Seq data of lentiviral transduced CD4 T cells can be accessed in the Gene Expression Omnibus (GEO) repository under the accession number GSE214888. The ChIP-Seq and RNA-Seq data of B-EBV cells can be accessed in the GEO repository under the accession numbers GSE199684 and GSE199685 (GSE199686 SuperSeries).