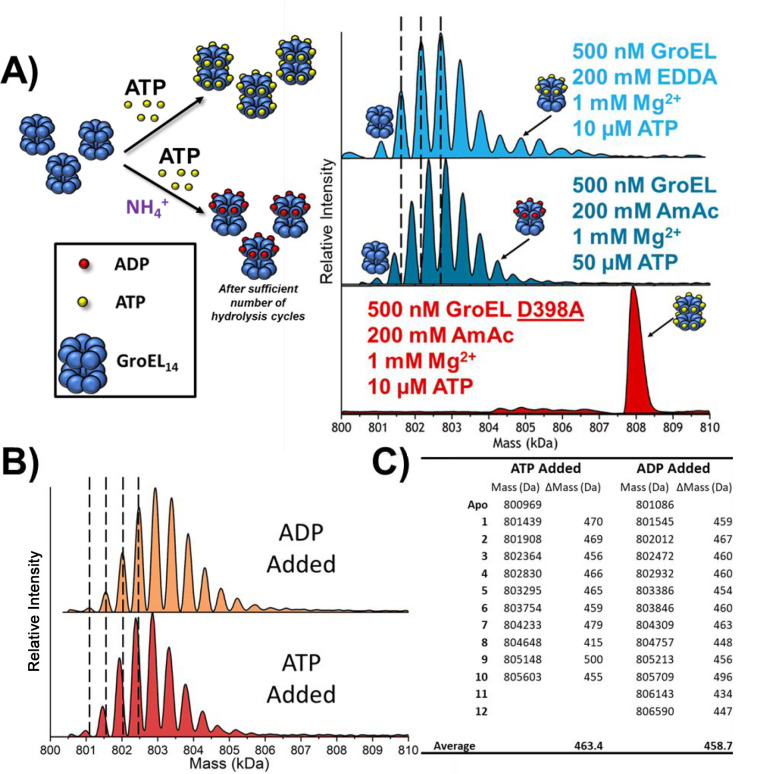
Figure 3.
(A) Stacked spectra showing the difference in mass shifts for nucleotide binding observed in EDDA and AmAc solutions. Black lines are used to aid the viewer and show that the mass shifts for EDDA are assigned to [ATP + Mg2+]n while the mass shifts for AmAc conditions are assigned to [ADP + Mg2+]n. A hydrolysis-deficient mutant GroELD398A was also analyzed in an AmAc solution (red), which shows the elevated level of cooperative binding in the presence of NH4+ ions and the absence of hydrolysis. Also note for GroELD398A that the affinity and cooperativity of the GroEL mutant for ATP are drastically increased in AmAc compared to EDDA conditions. (B) Stacked deconvoluted mass spectra showing the similarities in the binding distributions when either ADP is directly added to a solution containing GroEL or ATP is added under conditions where hydrolysis occurs. Solution conditions are 500 nM GroEL, 50 μM ATP or ADP, 1 mM MgAc2, and 200 mM AmAc at 25 °C. (C) Table containing the peak centroid data for (A). Mass shift values are in the form ΔMass = (n + 1) – n. It should be noted that the measured mass of the apo complex for ATP vs ADP is shifted by about 120 Da, explaining why subsequent peaks are not exactly aligned in (B).