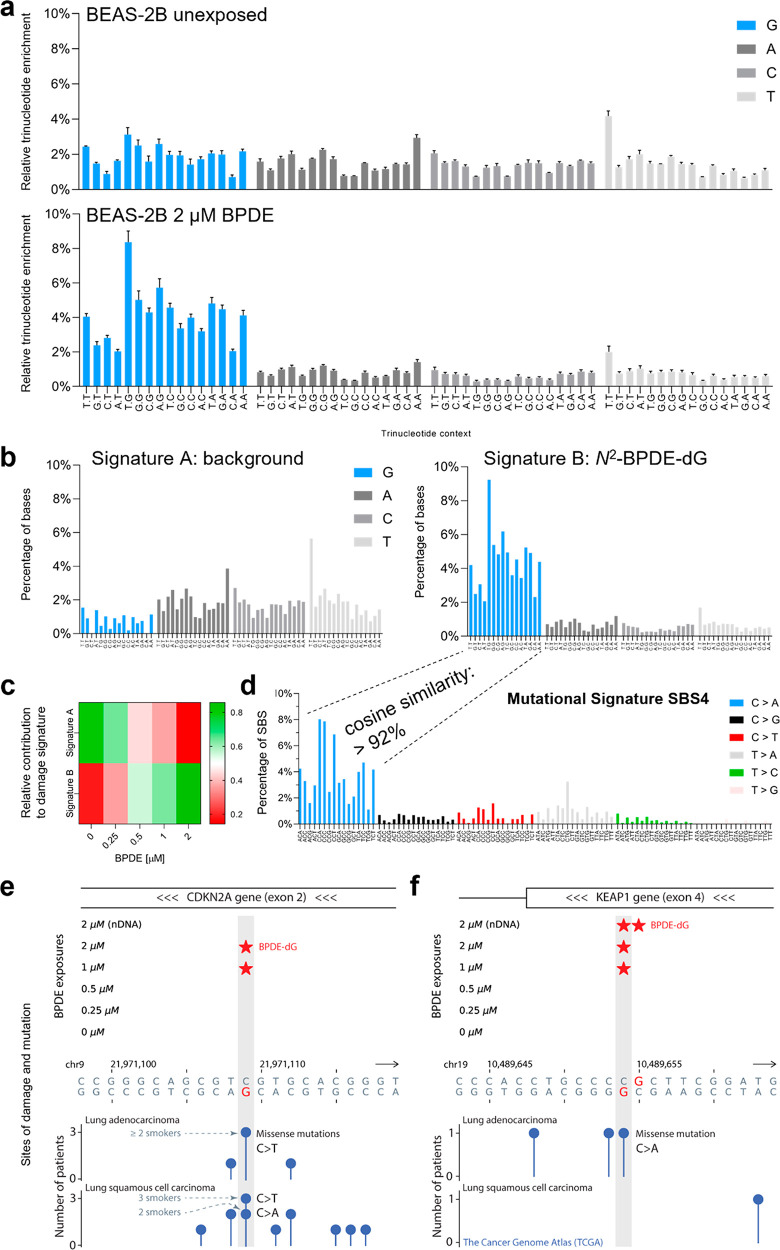
Figure 4.
(a) Relative trinucleotide enrichment at the predicted damage site from nonexposed cells and cells exposed to 2 μM BPDE. (b) Signatures extracted from all samples using a non-negative matrix factorization. Two signatures were extracted and labeled A and B. (c) Heat map representing the relative contribution of Signature A and B to the damage signature extracted from cells exposed to different BPDE concentrations. (d) Mutational Signature 4 extracted from smoking-related lung cancers. Cosine similarity of the extracted relative trinucleotide enrichment profile with C > A mutations in Signature 4. (e–f) BPDE-dG at lung-cancer-associated mutation sites located in tumor suppressor genes CDKN2A (e) and KEAP1 (f). A star indicates BPDE damage called by at least two reads across experimental replicates of a given condition. The identified damage-mutation match in CDKN2A is the most frequently mutated site of this gene in lung adenocarcinomas. As a reference for the Y-axis values of the mutation data in KEAP1, for ∼93% of lung-carcinoma mutation sites the number of patients with mutation is one, and for the remaining mutation sites this number is two. The TCGA mutational data were obtained via UCSC Table Browser. The gene annotation is according to GENCODE V41 (MANE-only set).