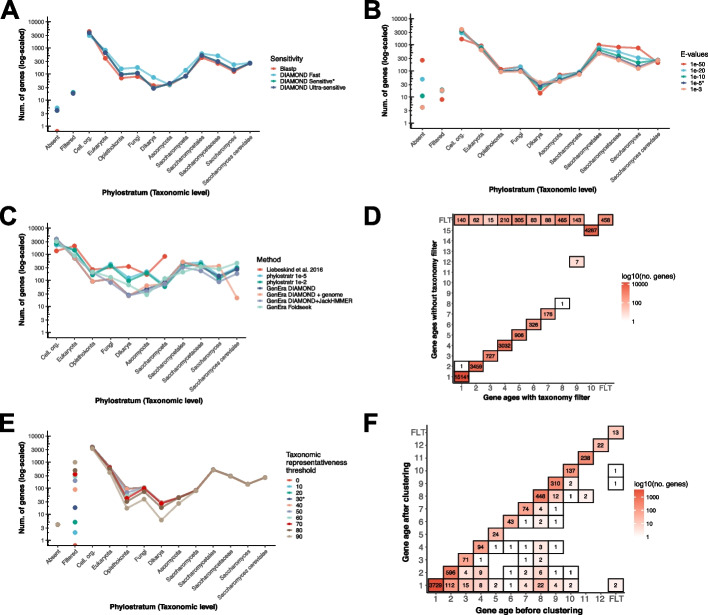
Fig. 3.
Benchmarking of GenEra through the analysis of Saccharomyces cerevisiae (A–C and E, F) and Apostichopus japonicus (D). A DIAMOND in ultra-sensitive and sensitive mode (*default parameter) generates a similar pattern of gene age assignment as the gold standard BLASTP while using the same e-value threshold of 10−5. The search sensitivity level does not influence the number of genes that are filtered through the taxonomic representativeness threshold (filtered) and has a negligible effect on the number of genes that fail to match themselves through pairwise alignment (absent). B The patterns of gene age assignment remain largely unaffected between a permissive e-value threshold of 10−3 and a more stringent threshold of 10−5 (*default parameter). Using more stringent thresholds (10−10 or lower) leads to an overrepresentation of TRGs at younger taxonomic levels. Lower e-value thresholds also increase the number of genes whose self-alignment cannot be detected (absent), thereby increasing the amount of false negative matches in the database. C GenEra can uncover deeper evolutionary relationships compared with previously published methods [24, 35], as seen in the number of genes that are traced back to the LUCA (cellular organisms). Using GenEra with additional 6-frame genome searches reduces the number of TRGs in the youngest taxonomic levels, from the species level up to the genus level, but older taxonomic levels are unaffected when including protein-against-genome data. Using JackHMMER increases the sensitivity at detecting homologs within older taxonomic levels, but shows little effect at finding homologs in the youngest taxonomic levels. Foldseek also increases the sensitivity at older levels but overestimates the number of genes at the species and genus levels. D Gene age assignments of Apostichopus japonicus before and after accounting for taxonomic levels lacking complete genomic data. The incomplete sampling of genomes across different taxonomic levels hinders gene age assignments, such as artificial patterns of gene absence that are erroneously filtered as contamination or HGT events (FLT). We established a parameter to exclude the taxonomic levels lacking genomic data, which improves the assignment of gene ages. E Taxonomic representativeness thresholds have a direct impact on the number of genes that can be assigned to a specific age (filtered). We established a default threshold of *30%, as lower values are bound to represent artifacts due to genome contamination and false positive matches while more stringent thresholds fail to account for gene losses and incomplete genome databases. F The clustering step helps to track down the founder events of some genes with limited traceability that share a common founder event with other paralogs of the same gene family, which is reflected in older gene age assignments