Figure 5: Ester and aspartimide formation in amycolimiditide variants.
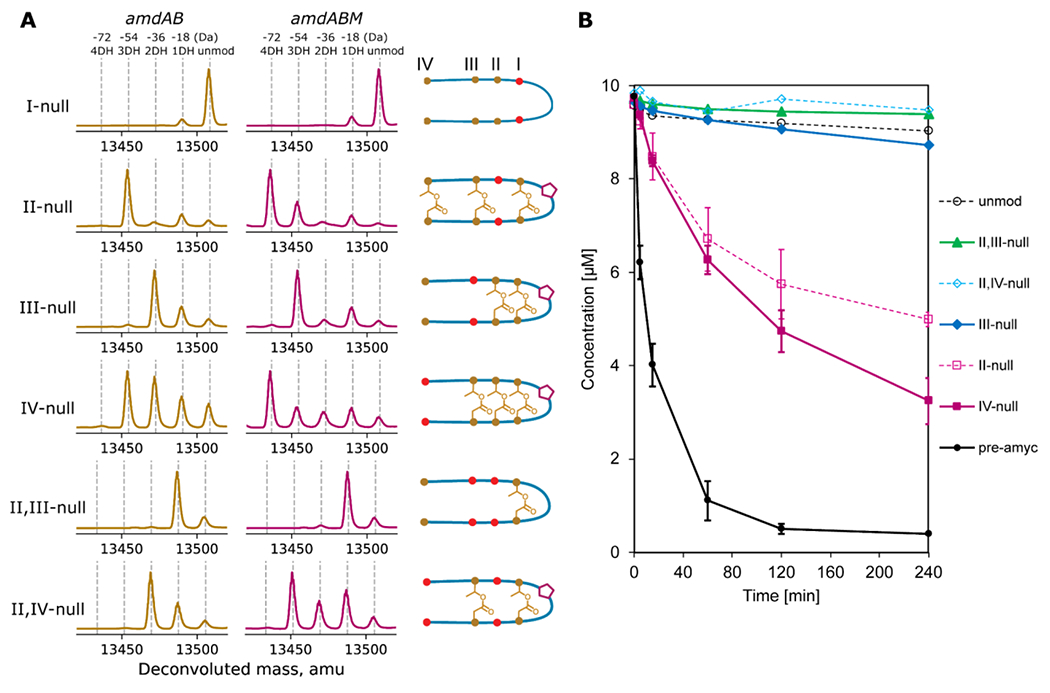
A: Variants of amycolimiditide with different esters disrupted were coexpressed with either AmdB or AmdB and AmdM. The notation I-null indicates that both residues involved in forming the I-ester (T10 and D21) have been substituted with isosteric but non-reactive residues (T10V and D21N). The positions of these substitutions are denoted by red dots in the cartoon, which represent the most probable structure of the variants. The deconvoluted mass spectra in the amdAB column show the number of dehydrations upon coexpression with AmdB while the cartoons for each variant show the position of the esters as determined by hydrazinolysis (Fig. S31–35). The mass spectra in the amdABM column show the number of dehydrations for each of the variants upon coexpression with both AmdB and AmdM. The cartoons also show whether the variants are aspartimidylated by AmdM (pentagon) B: Time course of in vitro aspartimidylation of pre-amycolimiditide (pre-amyc), unmodified core peptide (unmod), and variants. Disruption of any esters, even a distal one like in the IV-null case, has a large negative effect on the rate of aspartimidylation. The experiments on pre-amyc, II-null, and IV-null were repeated in triplicate and error bars reflect the standard deviation.
