Figure 7: Summary of amycolimiditide biosynthesis. A: Model of amycolimiditide biosynthesis.
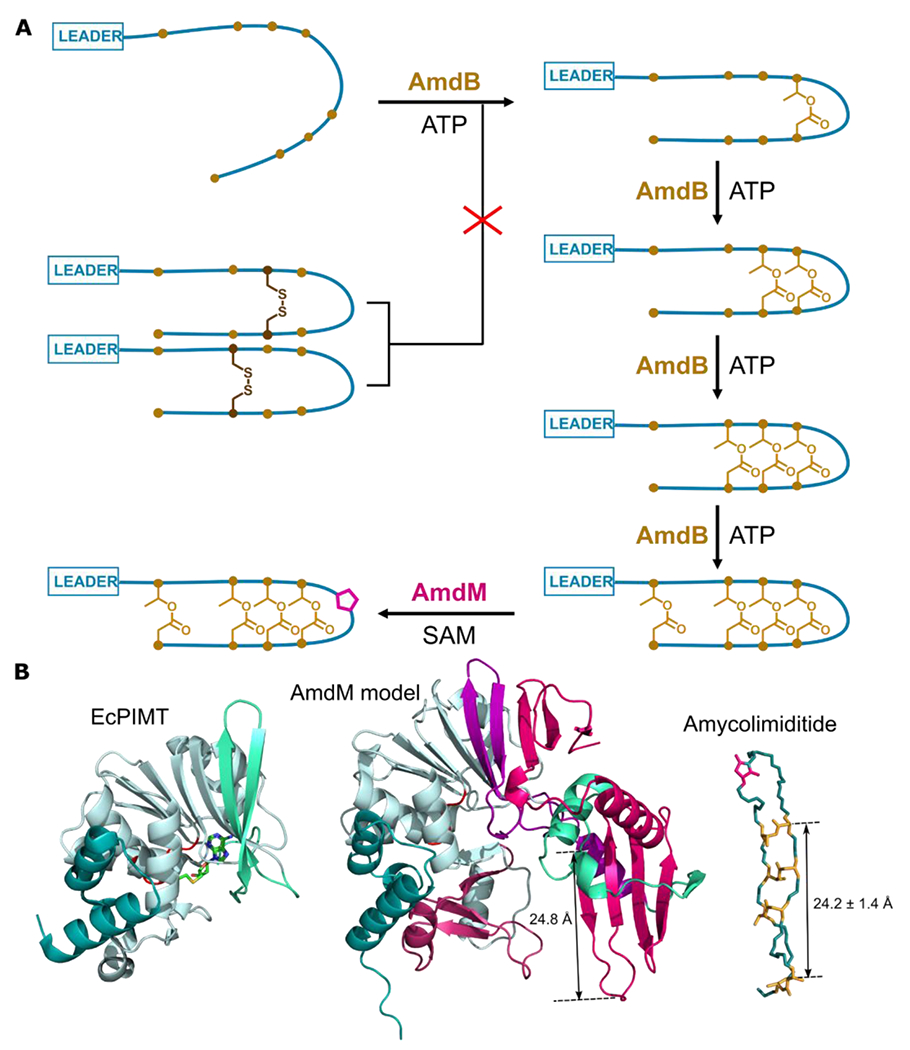
The esters in the amycolimiditide form prior to aspartimidylation. Preconstraining the substrate with disulfide bonds prevents ester formation, suggesting that the substrate must be flexible in order to be recognized by the ATP-grasp enzyme AmdB. The esters are installed in an ordered fashion, starting from the loop and proceeding down the stem. The four-fold esterified product is most efficiently recognized and methylated by the O-methyltransferase AmdM to give the final aspartimidylated graspetide. B: Comparison of E. coli PIMT (EcPIMT, PDB code 3LBF, left) to AlphaFold model of AmdM (middle). Homologous sequence segments are colored the same; see also Fig. S43 for an alignment of these two proteins. A molecule of S-adenosyl homocysteine (SAH) is bound in the methylation active site of EcPIMT. The model of AmdM includes a large C-terminal extension relative to EcPIMT (magenta) that generates a potential substrate binding cleft within the enzyme. The length of a β-strand that may act as a substrate recognition site matches closely the length of amycolimiditide shown on the right.
