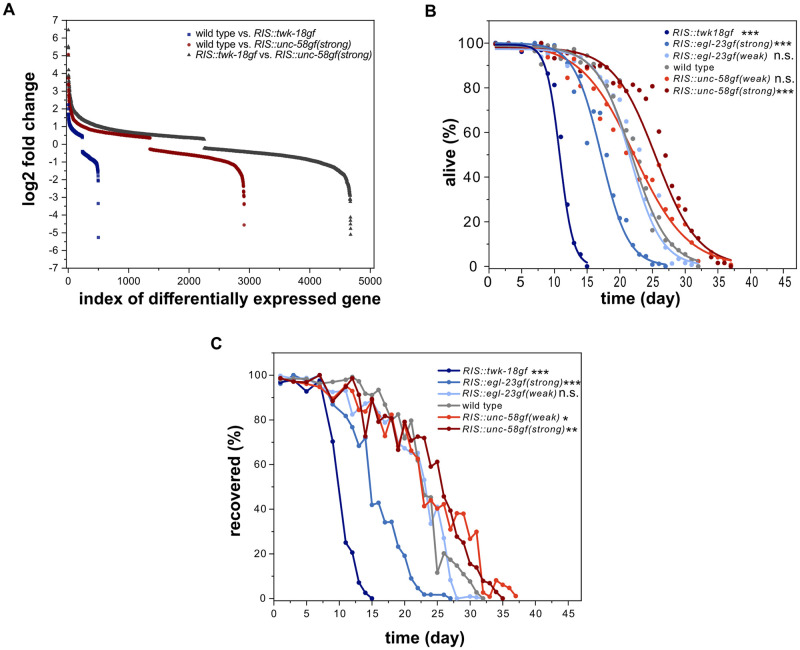
Fig 2. Survival of starvation-induced L1 arrest is a function of RIS activity.
A) RIS::twk-18gf and RIS::unc-58gf(strong) strains cause divergent transcriptional profiles. Shown is the log2 fold change for all differentially expressed genes. A gene was counted as differentially expressed with an FDR<0.05. According to our previously published results, RIS::twk-18gf expressed 498 genes differentially compared with the wild type. RIS::twk-18gf has been shown to be similar to aptf-1(gk794) [24]. RIS::unc-58gf(strong) was shown to express 2913 genes differentially compared with the wild type [24]. Comparing RIS::twk-18gf and RIS::unc-58gf(strong) revealed 4676 differentially expressed genes. This analysis supports the view that these two strains have opposing transcriptional profiles and identifies 1910 additional genes that are potentially under the control of RIS activity (S5 Table). B) Strong inactivation significantly decreases survival in L1 arrest whereas strong activation prolongs survival (three-parameter logistic fit, see also S1 Table, Fisher’s Exact Test was conducted when 50% of wild-type worms were alive (day 22)). The p-values were FDR corrected with Benjamini-Hochberg procedure with a 5% false discovery rate. The plot includes data from three replicates. ***p<0.001. C) Recovery from L1 arrest depends on RIS activity with strong inactivation leading to a reduced recovery and constant activation leading to a prolonged ability to recover. For statistics, the Fisher’s Exact Test was conducted at the time when 50% of wild-type worms recovered (day 23). The p-values were FDR corrected with Benjamini-Hochberg procedure with a 5% false discovery rate. The plot includes data from two replicates. N.s. p>0.05, *p<0.05, **p<0.01, ***p<0.001. Fisher’s Exact Test.