Abstract
Aims
Current non-invasive screening methods for cardiac allograft rejection have shown limited discrimination and are yet to be broadly integrated into heart transplant care. Given electrocardiogram (ECG) changes have been reported with severe cardiac allograft rejection, this study aimed to develop a deep-learning model, a form of artificial intelligence, to detect allograft rejection using the 12-lead ECG (AI-ECG).
Methods and results
Heart transplant recipients were identified across three Mayo Clinic sites between 1998 and 2021. Twelve-lead digital ECG data and endomyocardial biopsy results were extracted from medical records. Allograft rejection was defined as moderate or severe acute cellular rejection (ACR) based on International Society for Heart and Lung Transplantation guidelines. The extracted data (7590 unique ECG-biopsy pairs, belonging to 1427 patients) was partitioned into training (80%), validation (10%), and test sets (10%) such that each patient was included in only one partition. Model performance metrics were based on the test set (n = 140 patients; 758 ECG-biopsy pairs). The AI-ECG detected ACR with an area under the receiver operating curve (AUC) of 0.84 [95% confidence interval (CI): 0.78–0.90] and 95% (19/20; 95% CI: 75–100%) sensitivity. A prospective proof-of-concept screening study (n = 56; 97 ECG-biopsy pairs) showed the AI-ECG detected ACR with AUC = 0.78 (95% CI: 0.61–0.96) and 100% (2/2; 95% CI: 16–100%) sensitivity.
Conclusion
An AI-ECG model is effective for detection of moderate-to-severe ACR in heart transplant recipients. Our findings could improve transplant care by providing a rapid, non-invasive, and potentially remote screening option for cardiac allograft function.
Keywords: Artificial intelligence, Cardiac allograft rejection, Deep learning, Electrocardiography, Heart transplantation
Graphical Abstract
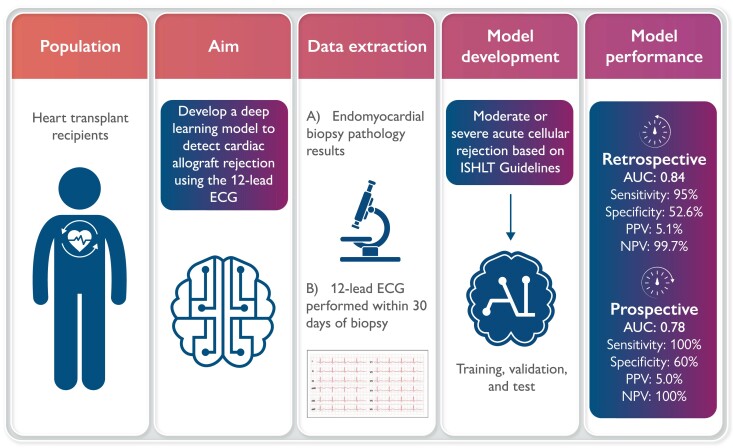
Graphical abstract.
An artificial intelligence enabled ECG can effectively detect cardiac allograft rejection among heart transplant recipients. AI, artificial intelligence; AUC, area under the receiver operating characteristic curve; ECG, electrocardiogram; ISHLT, International Society for Heart and Lung Transplantation; NPV, negative predictive value; PPV, positive predictive value.
Introduction
Current clinical practice at some centres following heart transplantation still rely on multiple endomyocardial biopsies (EMBs)1,2 every 1–4 weeks within the first year (each patient having up to 14 biopsies or more) to screen for allograft rejection.3,4 Beyond the inconvenience related to frequent hospital visits and the associated cost of an EMB procedure, complications can also occur from EMBs.5 These can range from mild, such as pericardial effusion, to severe as seen with significant tricuspid valve regurgitation requiring repeat cardiac surgery. As such, it is imperative that non-invasive, safer, options are made available to heart transplant recipients to screen for cardiac allograft rejection. Developing novel diagnostic tools that may allow for remote screening would improve our ability to monitor patients with increased frequency and reduce the need for frequent visits to the transplant centre for all their care.
Over the past decade, less-invasive options to screen for cardiac allograft rejection have been developed to address this need. While some centres have taken advantage of these novel methods to reduce the number of EMBs performed, there remains significant variability across heart transplant centres regarding the use of these novel methods and number of EMBs performed at each site within the first year of heart transplantation. These changes in practice highlight the transplant community’s desire to find non-invasive methods to detect allograft rejection. These methods include the use of gene expression profiling6 (GEP) and donor-derived cell-free DNA (dd-cfDNA),7,8 with both methods requiring a blood draw.9 However, studies utilizing these methods have shown limited discrimination6–8 to detection of acute cellular rejection (ACR).
Electrocardiogram (ECG) changes are known to occur following a cardiac transplant, including a completely new ECG signature (donor heart) different from the native/recipient heart as well as changes related to both sympathetic denervation and reinnervation such as increased heart rate and QT changes.10–12 Some ECG changes have also been reported at the time of severe cardiac allograft rejection including low voltages and conduction abnormalities.13–16 It is unknown if subtle changes on the ECG correlating with cardiac allograft rejection can be detected using machine-learning methods. Our objective was to develop and validate a deep-learning model, leveraging the effectiveness of digital ECG signals to provide a scalable framework for non-invasive evaluation of cardiac allograft rejection in heart transplant recipients. Thus, providing a means to evaluate cardiac allograft function in clinical and non-clinical settings, provide remote care, and potentially reduce the need for EMBs among patients with a low likelihood of allograft rejection.
Methods
Study design
We initially conducted a retrospective cohort study to evaluate the ability of an artificial intelligence model to identify cardiac allograft rejection using data from a 12-lead ECG (AI-ECG). Following model development, we initiated a proof-of-concept prospective single-arm observational study to validate the effectiveness of the developed model for detection of allograft rejection.
Study population
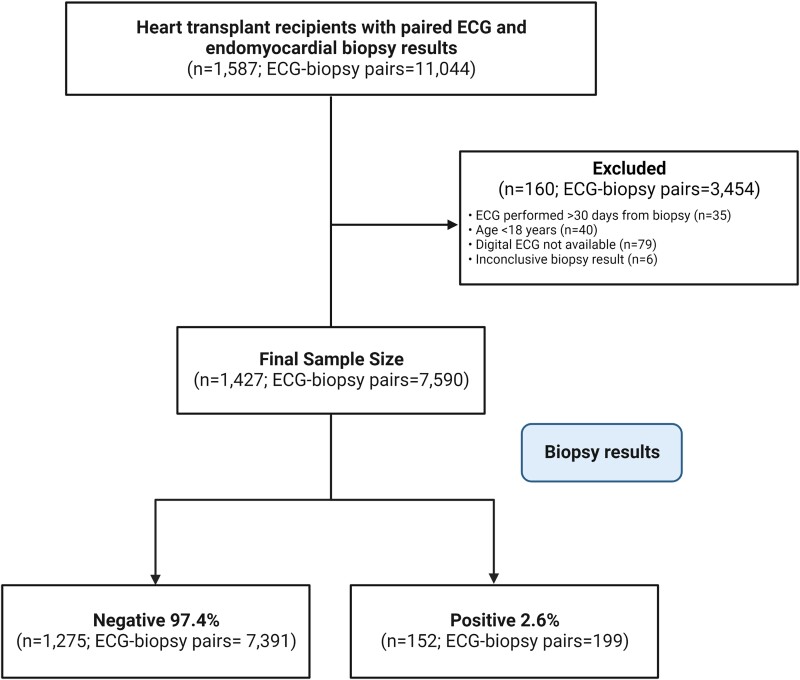
We retrospectively identified 1587 unique patients who had a heart transplant at three Mayo Clinic US sites between January 1988 and April 2021 with 11 044 unique ECGs performed within 30 days prior to an EMB. All EMBs were performed at the discretion of the ordering providers either as part of routine rejection surveillance or for any other clinical indication. We then limited the data set to unique ECGs matched to the closest EMB performed within 30 days prior. Only one ECG was selected for each EMB pathology result. We excluded patients younger than 18 years, those without digital ECGs and those with inconclusive EMB pathology results. Our final study sample corresponded to 1427 unique heart transplant recipients and 7590 unique ECG-biopsy pairs for model training and development (Figure 1). A large proportion (72.4%, n = 5495) of ECGs were performed within 7 days prior to EMB.
Figure 1.
Study flow diagram.
In recognition of the need for further validation studies, we designed a proof-of-concept screening study to prospectively gather ECGs in order to provide a preliminary estimate of model performance in a setting that might ultimately mirror a clinical implementation of the algorithm. We enrolled 56 consecutive heart transplant recipients following informed consent at Mayo Clinic in Florida, between 19 October and 8 December 2021, with 97 unique ECG-biopsy pairs. The criteria for inclusion in the study were: age 18 years and older, heart transplant recipient, and scheduled for an EMB. Study exclusions were age <18 years or unable to provide informed consent. Standard 12-lead ECGs were performed in the supine position on the same day prior to a scheduled EMB procedure, as such EMB pathology results were not available to study staff at the time of ECG recording. EMB procedures were performed, and results reported according to standard clinical protocols by the managing provider. AI-ECG prediction results were not made available to the proceduralist or pathologist prior to the EMB procedure and result reporting.
Measures
We extracted all standard 12-lead ECG digital data and EMB results for the study cohort from the electronic health record. All ECGs were acquired at a sampling rate of 500 Hz using a GE Marquette ECG machine (Marquette, WI, USA) and stored using the MUSE ECG data management system (GE Healthcare, Chicago, IL, USA). Cardiac allograft rejection was defined as moderate or severe ACR based on the International Society for Heart and Lung Transplantation (ISHLT) guidelines from 1990, 2004, and 2013.17,18 Antibody-mediated rejection (AMR) was not included due to variations in its definition, changes in key elements required for diagnosis over the years, and lack of routine screening in many heart transplant centers.19 The EMB results were extracted using a text-processing algorithm to identify ACR classified as none or mild (ACR negative) and moderate or severe (ACR positive). None or mild rejection included ISHLT 1990 Grades 0R, 1A, 1B, and 2 and ISHLT 2004 Grades 0R and 1R. Moderate or severe rejection included ISHLT 1990 Grades 3A, 3B, and 4 and ISHLT 2004 Grades 2R and 3R. The final classification groups were reviewed and approved by a cardiovascular pathologist and transplant cardiologist. Demographic and other clinical variables were abstracted from the electronic health records through a unified data platform. Co-morbid conditions were classified into disease groups using International Classification of Diseases (ICD) 9 and 10 diagnosis codes.20,21 Co-morbid conditions were included if the diagnosis was present in the medical records after 60 days following heart transplant in order to exclude post-operative complications that are expected to resolve.
Model development
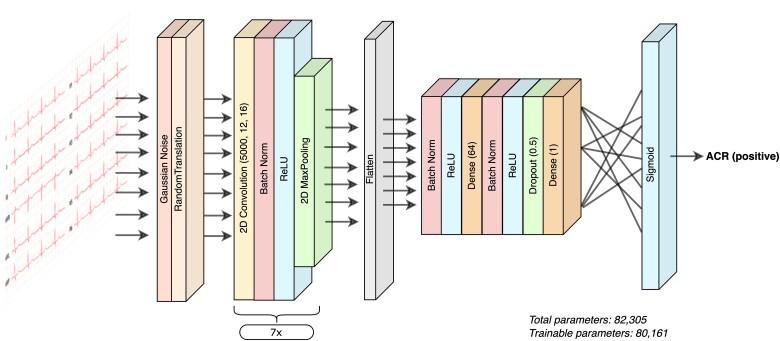
Our retrospective data set was split into three sets: training (80%), validation (10%), and test (10%). All samples from the same patient were kept within a specific set. The training/validation/test sets contained 1146/141/140 unique patients and 6074/758/758 unique ECG-biopsy pairs, respectively. Of those, 127/14/11 unique patients with 159/20/20 ECG-biopsy pairs were positive for ACR. A convolutional neural-network model was trained using the Keras framework with Tensorflow (Google, Mountain View, CA, USA) and backend implemented in Python,22 using the 12-lead ECG signal as input. The AI-ECG model consisted of 39 layers, with 80 161 trainable parameters. The input to the model is a 12 by 5000 array, representing each of the 12 leads for 10 s at 500 Hz. The first two layers apply random noise and translation to the ECG arrays, followed by repeated blocks of convolution, batch normalization, activation, and pooling. The final layers consisted of a dense sigmoid activation function, whose output represents the probability of ACR (Figure 2). The model was trained using binary cross-entropy as the loss function on the training set, after which, the predictions were obtained on the validation set, and the area under the receiver operating curve (AUC) and other diagnostic performance metrics were computed. The best model was chosen after a randomized hyperparameter search over optimizer, learning rates, class weights, batch size, and regularization. Final model performance was assessed on the test data set.
Figure 2.
Artificial intelligence electrocardiogram—model architecture.
Statistical analysis
Sample demographic and clinical characteristics data were summarized at the patient level with median, lower quartile (Q1), and upper quartile (Q3) for the continuous variables and with frequencies and percentages for the categorical variables. ECG characteristics were similarly summarized for each available ECG. The Wilcoxon rank sum test for continuous variables and Fisher’s exact test for categorical variables were used to compare differences in demographic and clinical characteristics between patients classified as positive or negative for ACR.
For diagnostic performance of the AI-ECG model, the reported measures of diagnostic accuracy were based on the test data. There were two levels of clustering present in the data. At the highest level, patients had multiple biopsies. Then for each biopsy, a lower level of clustering could have occurred when multiple ECGs were obtained prior to EMB results. To account for this statistically, the analyses selected the ECG obtained most recently prior to the EMB procedure to remove the lower level clustering from the data. For the higher level of clustering in the data, the analysis assumed statistical independence of the repeated EMBs within a patient. This decision, while subject to some statistical limitations [confidence intervals (CIs) that are more precise due to not accounting for the design effect imposed by the clustering], mimics how the algorithm would be utilized in clinical practice. To assess the sensitivity of this limitation, a patient-level analysis using only the last EMB-ECG pairing was also evaluated.
A receiver operating characteristic (ROC) curve was generated in the retrospective test data set by computing the predicted probabilities from the neural-network model and evaluating the true-positive rate and false-positive rate for various threshold values ranging from 0 to 1. Sensitivity, specificity, positive predictive value, and negative predictive value were calculated, with each ECG-biopsy pair, treated as an independent unit, using a threshold predicted probability value of 0.4, selected as the optimal threshold to maximize the sensitivity of the test relative to specificity.
For the prospective study, we modelled the prediction probabilities for ACR generated by the developed AI-ECG model in relation to EMB pathology results for rejection using ECGs performed on the same day as the EMB procedure. Measures of diagnostic performance based on dichotomized predictions were obtained at the previously determined threshold of ≥0.4 to indicate a positive AI-ECG screen. CIs were also calculated for all measures of diagnostic performance using exact methods. For the AUC, the large sample approximation of the DeLong method23 with optimization by Sun and Xu24 was used. All statistical analyses were performed using R version 4.0.3 (Vienna, Austria)25 and a P-value of <0.05 was considered statistically significant.
Results
Sample clinical and ECG characteristics are provided in Tables 1 and 2. The median age at last biopsy in the retrospective cohort was 58.3 years (Q1: 48.7, Q3: 65.2) and 28.9% were females. The median number of ECG-biopsy pairs per patient was 5 (Q1: 2, Q3: 7). The majority (97.4%) of EMB results were classified as none or mild rejection and 2.6% had moderate or severe rejection. Heart transplant recipients who had at least one episode of ACR were more likely to be younger (P < 0.001) and further out from their transplant date (P < 0.001). They were also more likely to have a documented diagnosis of any cancer (P = 0.022) following heart transplant. There were also notable differences in ECG features obtained during an episode of ACR (Table 2) including higher heart rate, longer PR, QRS, and corrected QT intervals. ACR-positive ECGs were also less likely to be in sinus rhythm (P = 0.003), whereas sinus tachycardia (P = 0.034), right bundle branch block (P = 0.046), and atrial fibrillation (P = 0.015) were more prevalent. Given, the observed differences, we also evaluated the ability of the ECG variables to detect ACR and performance was inferior to the AI-ECG model with AUC value of 0.579 (see Supplementary material online, Figure S1).
Table 1.
Demographic and clinical characteristics of heart transplant recipients by cardiac allograft rejection status in the retrospective cohort
| Median (Q1, Q3) or no. (%) of patients | |||||
|---|---|---|---|---|---|
| Characteristic | n | Overall (n = 1427) | ACR (positive) (n = 152) | ACR (negative) (n = 1275) | P-value |
| Demographics | |||||
| Age at heart transplant (years) | 1427 | 55.6 (45.1, 62.0) | 52.1 (39.1, 58.5) | 56.0 (45.8, 62.2) | <0.001 |
| Age at last biopsy (years) | 1427 | 58.3 (48.7, 65.2) | 56.5 (46.6, 64.5) | 58.4 (49.0, 65.4) | 0.096 |
| Sex (female) | 1427 | 413 (28.9%) | 54 (35.5%) | 359 (28.2%) | 0.072 |
| Time since heart transplant to last biopsy (months) | 1427 | 24.9 (5.1, 59.1) | 49.4 (23.7, 91.2) | 24.2 (3.8, 49.9) | <0.001 |
| Race | 1402 | 0.45 | |||
| ȃBlack or African American | 179 (12.8%) | 18 (12.2%) | 161 (12.8%) | ||
| ȃOther | 81 (5.8%) | 5 (3.4%) | 76 (6.1%) | ||
| ȃWhite | 1142 (81.5%) | 124 (84.4%) | 1018 (81.1%) | ||
| Ethnicity (Hispanic or Latino) | 1272 | 108 (8.5%) | 8 (7.1%) | 100 (8.6%) | 0.72 |
| Echocardiographic features | |||||
| ȃLeft ventricular ejection fraction (%) | 1374 | 62.0 (58.0, 66.0) | 62.0 (58.0, 65.5) | 62.0 (58.0, 66.0) | 0.88 |
| ȃLeft ventricular end diastolic dimension (mm) | 1200 | 44.0 (41.0, 47.0) | 44.0 (41.0, 47.0) | 44.0 (41.0, 47.0) | 0.84 |
| ȃCardiac output (L/min) | 966 | 5.8 (5.0, 6.8) | 5.8 (5.1, 6.5) | 5.8 (5.0, 6.9) | 0.70 |
| Co-morbid conditions post-transplant | |||||
| ȃHypertension | 1418 | 1294 (91.3%) | 140 (92.7%) | 1154 (91.1%) | 0.65 |
| ȃDiabetes | 1418 | 799 (56.3%) | 75 (49.7%) | 724 (57.1%) | 0.083 |
| ȃPeripheral vascular disease | 1418 | 692 (48.8%) | 73 (48.3%) | 619 (48.9%) | 0.93 |
| ȃCerebrovascular disease | 1418 | 351 (24.8%) | 43 (28.5%) | 308 (24.3%) | 0.27 |
| ȃCancer | 1418 | 274 (19.3%) | 40 (26.5%) | 234 (18.5%) | 0.022 |
| ȃConnective tissue disease or rheumatic disease | 1418 | 75 (5.3%) | 8 (5.3%) | 67 (5.3%) | 1.00 |
P-values result from a Wilcoxon rank sum test (continuous variables) or Fisher’s exact test (categorical variables). The bold numeric values in the ‘P-value' column represent statistically significant results (i.e. P-value < 0.05). The bold values in the ‘characteristic' column represent group headings for example age, sex, time since, and race/ethnicity are all considered demographic variables.
Table 2.
Electrocardiogram characteristics of heart transplant recipients by cardiac allograft rejection status in the retrospective cohort
| Median (Q1, Q3) or no. (%) of patients | |||||
|---|---|---|---|---|---|
| Characteristic | n | Overall (n = 7590) | ACR (positive) (n = 199) | ACR (negative) (n = 7391) | P-value |
| Heart rate | 7572 | 92 (84, 103) | 95 (87, 106) | 92 (83, 103) | 0.009 |
| PR interval (ms) | 7526 | 148 (132, 168) | 156 (134, 170) | 148 (132, 166) | 0.049 |
| QRS duration (ms) | 7572 | 100 (88, 128) | 108 (92, 133) | 100 (88, 128) | 0.002 |
| QT interval (ms) | 7572 | 372 (348, 398) | 376 (351, 404) | 372 (348, 398) | 0.15 |
| Corrected QT interval (ms) | 7572 | 460 (439, 483) | 473 (444, 496) | 460 (439, 483) | <0.001 |
| Sinus rhythm | 7572 | 5107 (67.4%) | 114 (57.3%) | 4993 (67.7%) | 0.003 |
| Sinus tachycardia | 7572 | 2007 (26.5%) | 66 (33.2%) | 1941 (26.3%) | 0.034 |
| Sinus bradycardia | 7572 | 38 (0.5%) | 3 (1.5%) | 35 (0.5%) | 0.077 |
| 1st degree AV block | 7572 | 303 (4.0%) | 13 (6.5%) | 290 (3.9%) | 0.094 |
| Mobitz Type 1 AV block | 7572 | 10 (0.1%) | 1 (0.5%) | 9 (0.1%) | 0.23 |
| Left bundle branch block | 7572 | 65 (0.9%) | 1 (0.5%) | 64 (0.9%) | 1.00 |
| Right bundle branch block | 7572 | 2939 (38.8%) | 91 (45.7%) | 2848 (38.6%) | 0.046 |
| Atrial fibrillation/flutter | 7572 | 218 (2.9%) | 12 (6.0%) | 206 (2.8%) | 0.015 |
| Pacemaker (ventricular pacing) | 7572 | 452 (6.0%) | 13 (6.5%) | 439 (6.0%) | 0.65 |
| Prolonged QT | 7572 | 515 (6.8%) | 18 (9.0%) | 497 (6.7%) | 0.20 |
| Non-specific interventricular conduction delay (IVCD) | 7572 | 229 (3.0%) | 9 (4.5%) | 220 (3.0%) | 0.20 |
P-values result from a Wilcoxon rank sum test (continuous variables) or Fisher’s exact test (categorical variables). The bold numeric values in the ‘P-value' column represent statistically significant results (i.e. P-value < 0.05). The bold values in the ‘characteristic' column represent group headings for example age, sex, time since, and race/ethnicity are all considered demographic variables.
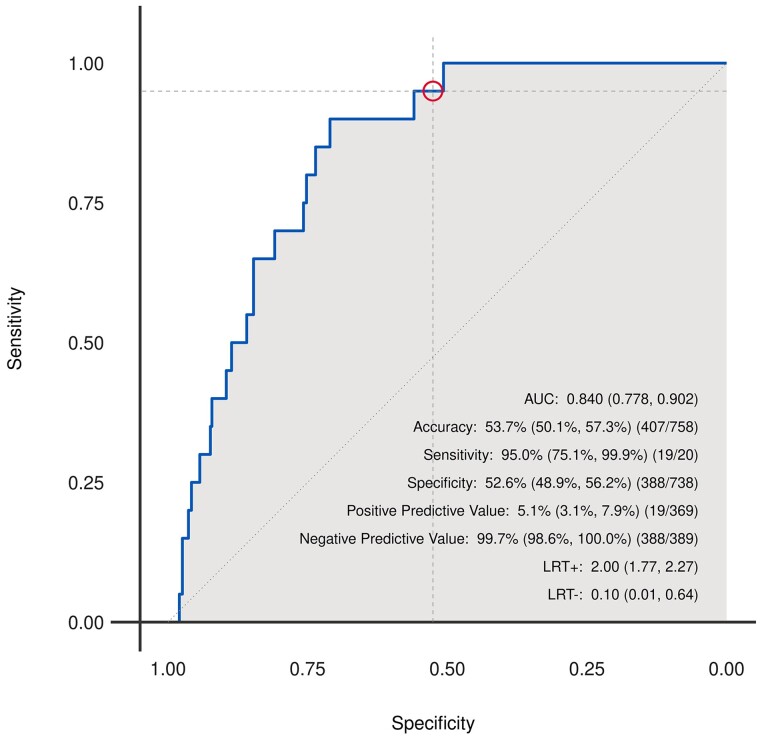
The AI-ECG model detected ACR with an AUC of 0.84 (95% CI: 0.78–0.90) in the test set (Figure 3). Additional metrics of diagnostic performance in the test set were sensitivity: 95% (95% CI: 75.1–99.9%; 19/20), specificity: 52.6% (95% CI: 48.9–56.2%; 388/738), positive predictive value: 5.1% (95% CI: 3.1–7.9%; 19/369), and negative predictive value: 99.7% (95% CI: 98.6–100%; 388/389).
Figure 3.
Receiver operating characteristic curve for detection of cardiac allograft rejection in a retrospective cohort of heart transplant recipients using a deep-learning model.
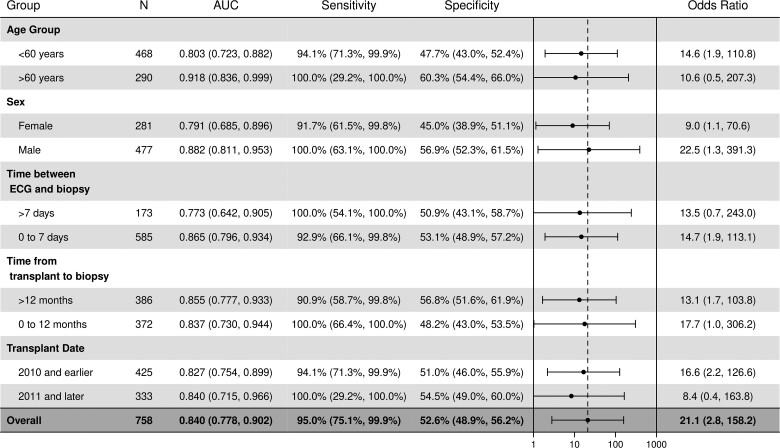
Subgroup analysis
We also evaluated model performance in specific patient subgroups. The subgroups included age, sex, time between ECG and biopsy, time from transplant to biopsy, and transplant date as shown in Figure 4. The AI-ECG model remained effective in each subgroup with slightly better performance among persons older than 60 years and men. Similar performance across subgroups was demonstrated in the training data set (see Supplementary material online, Figure S2). Given the model was trained using ECGs obtained within 30 days of an EMB, we considered different time points in the subgroup analysis. These were: ECGs obtained within 7 days and >7days of an EMB and compared model performance metrics across the time points. As shown in Figure 4 and Supplementary material online, Table S1, model performance was noted to be better for ECGs performed within 7 days of ECG (0.87) compared with >7 days (0.77). Similarly, we evaluated the AI-ECG model based on time since the heart transplant as follows: within 60 days (≤2 months), >60 days (>2 months), >90 days (>3 months), >120 days (>4 months), >180 days (>6 months), >365 days (>1 year), and >730 days (>2 years) days. The AUC values remained stable in these categories ranging from 0.81 to 0.89 (see Supplementary material online, Table S1). As described in the methods above, we also evaluated the performance of the model in a patient-level analysis using only the last EMB-ECG pairing and the AUC value in the training and test sets were 0.86 (95% CI: 0.80–0.93) and 0.87 (95% CI: 0.67–1.0) respectively.
Figure 4.
Subgroup analysis showing deep learning model performance in a retrospective cohort of heart transplant recipients.
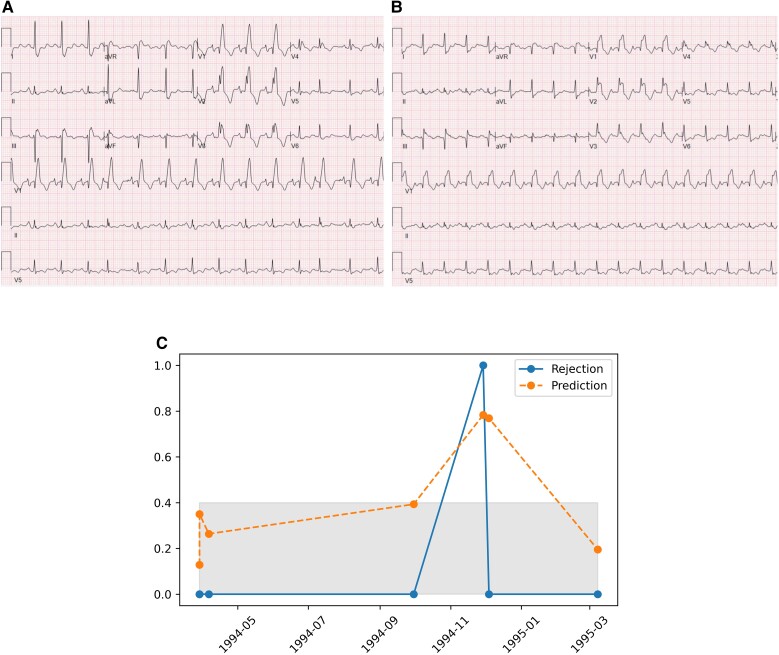
To demonstrate the unique ability of the AI-ECG model to detect ACR, we evaluated a patient example in the test data with multiple ECG-biopsy pairs and show an upward trend in AI-ECG prediction probabilities around an episode of ACR and how the prediction probability drops below the positive threshold following recovery from the episode of rejection (Figure 5). A review of the ECG images performed prior to and during the episode of ACR showed no clinically discernible changes (Figure 5).
Figure 5.
(A) Twelve-lead electrocardiogram obtained prior to rejection episode. (B) Twelve-lead electrocardiogram obtained during the rejection episode. (C) Deep learning prediction probabilities for a single patient over time—prior to, during an episode of, and following recovery from moderate acute cellular rejection. The grey-shaded area (corresponding to values less than 0.4) highlights prediction probabilities below the threshold for a positive screen.
Prospective proof-of-concept study
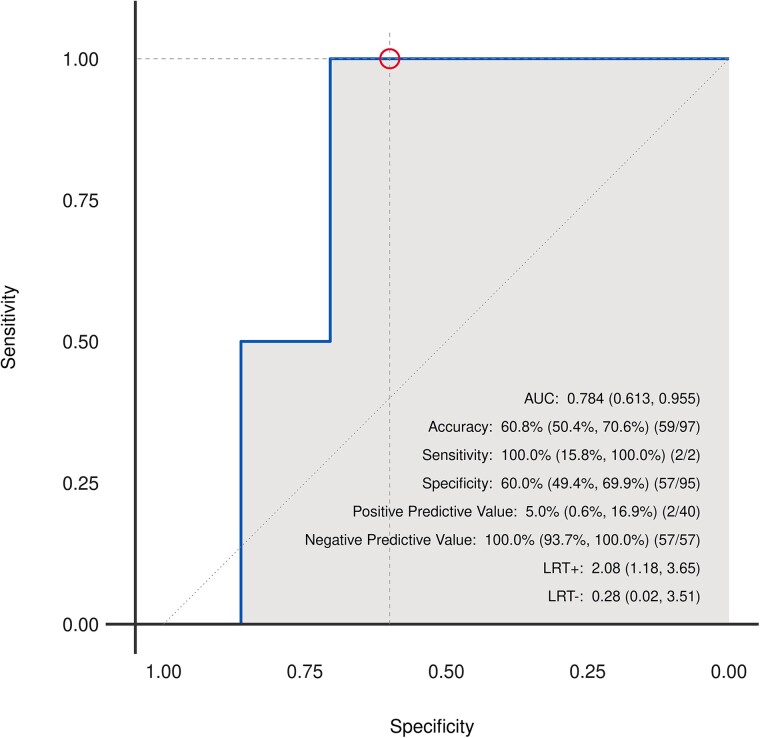
The prospective cohort included a total of 56 heart transplant recipients with 97 unique ECG-biopsy pairs. Sample clinical and ECG characteristics for the prospective cohort are provided in Supplementary material online, Tables S2 and S3. Approximately a quarter (26.8%) were females, and the median age at enrolment was 60.2 years (Q1: 55.5, Q3: 66.1). The developed AI-ECG model was able to identify ACR with an AUC of 0.78 (95% CI: 0.61–0.96; Figure 6). Sensitivity, specificity, positive predictive value, and negative predictive value were 100% (95% CI: 15.8–100%; 2/2), 60% (95% CI: 49.4–69.9%; 57/95), 5% (95% CI: 0.6–16.9%; 2/40), and 100% (95% CI: 93.7–100%; 57/57), respectively. All cases of ACR (2/2 ECG-biopsy pairs) were correctly identified and 34% (13/38 ECG-biopsy pairs) of the false-positive predictions were noted to have mild rejection based on EMB results. In addition, the two EMB confirmed positive ACR cases were noted to have had a positive AI-ECG prediction for ACR on analyses of ECGs performed ∼1 month prior, even though the EMB results at the time were negative for ACR. This suggests that the AI-ECG may potentially recognize subclinical ACR prior to confirmation with an EMB.
Figure 6.
Receiver operating characteristic curve for detection of cardiac allograft rejection in a prospective cohort of heart transplant recipients using a deep-learning model.
Exploratory analyses to estimate clinical use cases
The prior results represent the development and validation of the algorithm. These summaries do not capture how the algorithm might be used in the practice. To provide some initial data to inform the design of future studies examining the AI-ECG implementation, a set of exploratory analyses were also conducted. Supplementary material online, Figure S3 demonstrates a simulated use case based on the retrospective test data set. This analysis that assumes this tool would be used as the first-line screening test for ACR among heart transplant recipients. This analysis shows that ECG-based screening would result in 369 positive AI-ECG predictions, each of which would likely still go on to have confirmatory ACR testing. However, it could potentially reduce the need for a biopsy in more than half (51.3%, n = 389) of our patient cohort which may result in reduced health care utilization and cost. The percentage of cases screened as test negative, which is the main driver in this calculation, is directly affected by the selected threshold. While the initial threshold of 0.4 was selected to maximize the sensitivity of the test relative to specificity, alternative thresholds that maximize negative predictive value may also be of interest to rule out the need for biopsy based on a negative test result. In multiple simulations, as shown in Supplementary material online, Table S4 using an alternative threshold of 0.35 will yield a 100% sensitivity and 100% negative predictive value in the retrospective test data set, will lead to a 41% (314/758) reduction in number biopsies performed.
Discussion
In this study, we demonstrate for the first time that an AI-ECG can be an effective tool for detecting ACR among heart transplant recipients with an AUC of 0.84, showing improved performance when compared with AUC values for available blood test-based options for detecting ACR (0.64–0.70).6–8 We also performed a small, prospective proof-of-concept study to evaluate the effectiveness of the AI-ECG model and it remained robust.
Given that ECG changes are known to occur with ACR, some published studies have attempted to use ECG characteristics and features to predict the presence of rejection. Published results from a systematic review of 17 studies demonstrated significant variability in the study designs evaluating ECG features in relation to rejection26 as well as performance and concluded that the only effective approach available for detecting rejection was an EMB. Doering et al.27 published a study protocol that aimed to evaluate the utility of remote ECG monitoring for detecting ACR, and subsequent results in a sample of 220 heart transplant recipients showed that increased QRS, QT, QTc, and PR intervals, right bundle branch block, and fascicular block were associated with moderate to severe ACR13 but no AUC was provided. This study also suggested the need for computerized algorithms utilizing ECG data for the detection of ACR. Similarly in our study, we found multiple ECG variables to be associated with ACR including heart rate, PR interval, QRS duration, QTc, sinus tachycardia, RBBB, and atrial fibrillation. Although we found a combination of these variables using a logistic regression model had a lower AUC for detecting ACR compared with the deep-learning model, it is possible that some of these ECG features may potentially be features utilized by the deep-learning model and further analysis geared towards model explainability might provide potential insights into this.
Existing non-invasive options for detection of ACR are based on blood tests, GEP and dd-cfDNA6–8,28 with AUCs ranging from 0.64 to 0.69. In a study with 63 patients and 132 samples the combination of both tests yielded an AUC of 0.78.8 Non-published results described by the manufacturer of these tests also list AUC values of 0.68, 0.77, and 0.81 for GEP, dd-cfDNA and a combination of both tests, respectively.29 Three small studies evaluating the utility of dd-cfDNA to detect all forms of cardiac allograft rejection (ACR and AMR combined) among 65, 171, and 223 patients showed an AUC of 0.83,30 0.9231 and 0.8632 respectively. The AUCs from these studies are notably higher than prior studies likely because the outcome of interest was different as these studies included detection of both ACR and AMR. Studies evaluating the detection of ACR alone, otherwise have shown lower AUC values compared with our AI-ECG model.6–8 These tests require a blood draw by trained personnel with specific blood collection equipment/kits and samples need to be kept at a low temperature prior to transporting to the laboratory for analysis to ensure test quality.33 In addition, there are concerns for potential RNA degradation for samples stored for greater than 24 h which may also affect test results.34 More recent advances have demonstrated the effectiveness of serum microRNAs (miRNA) to effectively detect allograft rejection among heart transplant recipients with validation AUC of 0.80 for detection of ≥Grade 2R ACR using a single miRNA, albeit based on a small sample size (212 serum samples paired with EMB results).35 A prior study had demonstrated that four separate miRNAs were able to detect all forms of cardiac allograft rejection (ACR + AMR) with AUC values ranging from 0.87 to 0.98 in an external validation sample of 53 heart transplant recipients.36
The ECG, however, is an ideal tool in this patient population given the relative frequency of obtaining an ECG for other clinical indications as part of the management of heart transplant recipients. The ECG is inexpensive, painless, non-invasive, and can be performed by hospital or clinic staff with minimal training, compared with blood test methods and the results can be made available within a few minutes following ECG acquisition as opposed to non-invasive blood test options utilizing GEP and dd-cfDNA which have a turnaround time ranging from 3 to 4 days.28 In addition, the use of ECG data may unlock the possibility for remote monitoring for ACR, potentially reducing health care costs, decreasing the need for patients to travel to heart transplant centres for frequent follow-up studies, and improving patient satisfaction. Other studies utilizing a machine learning approach for evaluation of ACR have mostly focused on an automated assessment of EMB slides37–39 due to the recognized limitation of conventional histologic analysis of EMB tissue slides3 and these have shown excellent results with AUCs ranging from 0.92 to 93.37,38 More importantly, these machine-learning methods may potentially address limitations related to EMB testing, particularly interobserver variability in the reported results based on the ISHLT criteria.40
Limitations of our study include a relatively small sample size used for model derivation (n = 1427) and prospective cohort (n = 56) which limits finer analysis of multiple subgroups, limited explainability of the AI model, a known shortcoming of deep-learning models, and the exclusion of AMR. In addition, the smaller proof-of-concept had only two cases of rejection, so the area under the ROC curve was a coarse step function with very low precision, i.e. the 95% CI spanned 0.61–0.96; virtually the entire range of plausible values. Numerically, the AUC in the proof-of-concept study was lower (AUC of 0.78) when compared with the test sample used to evaluate the initial model performance (AUC of 0.84); however, these differences are difficult to interpret given the low precision of the estimate and the limited number of events. These limitations notwithstanding, we did demonstrate the model correctly identified all cases of ACR in the prospective study and 34% of those flagged as false positive were noted to have mild rejection based on EMB results. A previously developed AI-ECG algorithm for detection of low left ventricular ejection fraction in an unselected patient sample noted that patients with false-positive predictions had a four-fold higher risk of developing a low ejection fraction in the future.41 As such, it would be important to understand if the false-positive predictions from our AI-ECG model may represent early markers of subclinical allograft dysfunction. Additional next steps will include continued follow-up of the patients with false-positive predictions. It is important to clarify that a positive screen based on the AI-ECG is not intended to dictate therapeutic intervention, rather it is intended to support the need for confirmatory testing with an EMB or alternate testing. A negative AI-ECG screen would be otherwise reassuring and in the absence of other clinical evidence of rejection would allow an EMB be deferred at the discretion of the managing provider.
Clearly, there are clinically identifiable ECG changes seen during an episode of ACR as shown in Table 2, but none of these changes may raise a red flag for rejection in clinical practice and are not specific for allograft rejection alone. Our hypothesis is that subtle changes in addition to those identified clinically are likely present on the ECG early in the clinical course of ACR and these may or may not be overtly manifest when interpreting the ECG clinically. Situations like this, is where deep-learning models often excel as demonstrated with other similar studies utilizing AI-ECG models for cardiovascular disease detection.41–43 In addition, subsequent validation studies and clinical trials have demonstrated AI-ECG model reliability and effectiveness.44–46 Strengths of this study include the use of a truly non-invasive modality, the ECG, to screen for ACR and a proof-of-concept prospective analysis of the derived AI-ECG model. Potential use cases for this technology will include the ability to increase ACR screening frequency with serial ECGs which is non-invasive with minimal patient discomfort and screening for ACR in non-transplant centres without the capacity to perform EMB. More importantly, essential steps needed prior to adopting this method in clinical practice includes external, multi-centre, validation of findings from this study, larger, robust prospective studies including clinical trials to evaluate its potential impact on clinical outcomes, as well as implementation studies.
Conclusion
We demonstrate that a deep-learning model based on the 12-lead ECG can effectively detect moderate-to-severe ACR among heart transplant recipients with a proof-of-concept prospective analysis showing promising results. This study adds to the growing body of evidence showing the potential ability for artificial intelligence models to enhance cardiovascular care. Future directions would include evaluating the potential benefit of combining AI-ECG screening with blood testing methods, evaluation of the AI-ECG for detecting ACR and AMR combined, as well as AI-based EMB slide analysis and interpretation.
Supplementary Material
Acknowledgements
The authors thank staff and members of the Digital Innovation Laboratory, Mayo Clinic, Florida and the Departments of Cardiovascular Medicine, Transplant Medicine, and Laboratory Medicine & Pathology at Mayo Clinic in Florida, Minnesota, and Arizona for their contributions in generating the data used.
Contributor Information
Demilade Adedinsewo, Department of Cardiovascular Medicine, Division of Cardiovascular Diseases, Mayo Clinic, 4500 San Pablo Rd, Jacksonville, FL 32224, USA.
Heather D Hardway, Department of Quantitative Health Sciences, Mayo Clinic, Jacksonville, FL, USA.
Andrea Carolina Morales-Lara, Department of Cardiovascular Medicine, Division of Cardiovascular Diseases, Mayo Clinic, 4500 San Pablo Rd, Jacksonville, FL 32224, USA.
Mikolaj A Wieczorek, Department of Quantitative Health Sciences, Mayo Clinic, Jacksonville, FL, USA.
Patrick W Johnson, Department of Quantitative Health Sciences, Mayo Clinic, Jacksonville, FL, USA.
Erika J Douglass, Department of Cardiovascular Medicine, Division of Cardiovascular Diseases, Mayo Clinic, 4500 San Pablo Rd, Jacksonville, FL 32224, USA.
Bryan J Dangott, Department of Laboratory Medicine and Pathology, Mayo Clinic, Jacksonville, FL, USA.
Raouf E Nakhleh, Department of Laboratory Medicine and Pathology, Mayo Clinic, Jacksonville, FL, USA.
Tathagat Narula, Department of Transplantation, Mayo Clinic, Jacksonville, FL, USA.
Parag C Patel, Department of Transplantation, Mayo Clinic, Jacksonville, FL, USA.
Rohan M Goswami, Department of Transplantation, Mayo Clinic, Jacksonville, FL, USA.
Melissa A Lyle, Department of Transplantation, Mayo Clinic, Jacksonville, FL, USA.
Alexander J Heckman, Department of Cardiovascular Medicine, Division of Cardiovascular Diseases, Mayo Clinic, 4500 San Pablo Rd, Jacksonville, FL 32224, USA.
Juan C Leoni-Moreno, Department of Transplantation, Mayo Clinic, Jacksonville, FL, USA.
D Eric Steidley, Department of Cardiovascular Medicine, Mayo Clinic, Phoenix, AZ, USA.
Reza Arsanjani, Department of Cardiovascular Medicine, Mayo Clinic, Phoenix, AZ, USA.
Brian Hardaway, Department of Cardiovascular Medicine, Mayo Clinic, Phoenix, AZ, USA.
Mohsin Abbas, Department of Cardiovascular Medicine, Mayo Clinic, Rochester, MN, USA.
Atta Behfar, Department of Cardiovascular Medicine, Mayo Clinic, Rochester, MN, USA.
Zachi I Attia, Department of Cardiovascular Medicine, Mayo Clinic, Rochester, MN, USA.
Francisco Lopez-Jimenez, Department of Cardiovascular Medicine, Mayo Clinic, Rochester, MN, USA.
Peter A Noseworthy, Department of Cardiovascular Medicine, Mayo Clinic, Rochester, MN, USA.
Paul Friedman, Department of Cardiovascular Medicine, Mayo Clinic, Rochester, MN, USA.
Rickey E Carter, Department of Quantitative Health Sciences, Mayo Clinic, Jacksonville, FL, USA.
Mohamad Yamani, Department of Cardiovascular Medicine, Division of Cardiovascular Diseases, Mayo Clinic, 4500 San Pablo Rd, Jacksonville, FL 32224, USA.
Supplementary material
Supplementary material is available at European Heart Journal – Digital Health.
Ethics statement
This study was approved by the Mayo Clinic Institutional Review Board (IRB). The retrospective component of the study was deemed exempt by the IRB with waiver of consent. Informed consent was obtained for all participants enrolled in the prospective component of this study.
Funding
This study was made possible by the Mayo Clinic clinical practice committee innovation funds. D.A. is supported by the Mayo Clinic Women's Health Research Center and the Mayo Clinic Building Interdisciplinary Research Careers in Women’s Health (BIRCWH) Program funded by the National Institutes of Health (NIH) [grant number K12 HD065987]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Data availability
The data underlying this article will be shared on reasonable request to the corresponding author. All requests for raw and analysed data and related materials, excluding programming code, will be reviewed by the Mayo Clinic legal department and Mayo Clinic Ventures to verify whether the request is subject to any intellectual property or confidentiality obligations. Requests for patient-related data not included in the paper will not be considered. Any data and materials that can be shared will be released via a Material Transfer Agreement.
References
- 1. Caves P, Billingham M, Stinson E, Shumway N. Serial transvenous biopsy of the transplanted human heart improved management of acute rejection episodes. Lancet 1974;303:821–826. [DOI] [PubMed] [Google Scholar]
- 2. Costanzo MR, Dipchand A, Starling R, Anderson A, Chan M, Desai S, et al. The International Society of Heart and Lung Transplantation guidelines for the care of heart transplant recipients. J Heart Lung Transplant 2010;29:914–956. [DOI] [PubMed] [Google Scholar]
- 3. Peyster EG, Madabhushi A, Margulies KB. Advanced morphologic analysis for diagnosing allograft rejection: the case of cardiac transplant rejection. Transplantation 2018;102:1230–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Seferović PM, Tsutsui H, McNamara DM, Ristić AD, Basso C, Bozkurt B, et al. Heart Failure Association, Heart Failure Society of America, and Japanese Heart Failure Society position statement on endomyocardial biopsy. J Card Fail 2021;27:727–743. [DOI] [PubMed] [Google Scholar]
- 5. Pham MX, Teuteberg JJ, Kfoury AG, Starling RC, Deng MC, Cappola TP, et al. Gene-expression profiling for rejection surveillance after cardiac transplantation. N Engl J Med 2010;362:1890–1900. [DOI] [PubMed] [Google Scholar]
- 6. Crespo-Leiro MG, Stypmann J, Schulz U, Zuckermann A, Mohacsi P, Bara C, et al. Clinical usefulness of gene-expression profile to rule out acute rejection after heart transplantation: CARGO II. Eur Heart J 2016;37:2591–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Khush KK, Patel J, Pinney S, Kao A, Alharethi R, DePasquale E, et al. Noninvasive detection of graft injury after heart transplant using donor-derived cell-free DNA: a prospective multicenter study. Am J Transplant 2019;19:2889–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Crespo-Leiro M, Zuckermann A, Stypmann J, Mohacsi P, Grskovic M, Beausang J, et al. Increased plasma levels of donor-derived cell-free DNA correlate with rejection in heart transplant recipients: the CARGO II multicenter trial. J Heart Lung Transplant 2015;34:S31–S32. [Google Scholar]
- 9. Deng MC. The evolution of patient-specific precision biomarkers to guide personalized heart-transplant care. Expert Rev Precis Med Drug Dev 2021;6:51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Awad M, Czer LSC, Hou M, Golshani SS, Goltche M, Robertis MD, et al. Early denervation and later reinnervation of the heart following cardiac transplantation: a review. J Am Heart Assoc 2016;5:e004070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Thajudeen A, Stecker EC, Shehata M, Patel J, Wang X, McAnulty JH, et al. Arrhythmias after heart transplantation: mechanisms and management. J Am Heart Assoc 2012;1:e001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Moore JP, Alejos JC, Perens G, Wong S, Shannon KM. The corrected QT interval before and after heart transplantation. Am J Cardiol 2009;104:596–601. [DOI] [PubMed] [Google Scholar]
- 13. Hickey KT, Sciacca RR, Chen B, Drew BJ, Pickham D, Carter EV, et al. Electrocardiographic correlates of acute allograft rejection among heart transplant recipients. Am J Crit Care 2018;27:145–150. [DOI] [PubMed] [Google Scholar]
- 14. Lacroix D, Kacet S, Savard P, Molin F, Dagano J, Pol A, et al. Signal-averaged electrocardiography and detection of heart transplant rejection: comparison of time and frequency domain analyses. J Am Coll Cardiol 1992;19:553–558. [DOI] [PubMed] [Google Scholar]
- 15. Irwin ED, Bianco RW, Clack R, Grehan J, Slovut DP, Nakhleh R, et al. Use of epicardial electrocardiogram for detecting cardiac allograft rejection. Ann Thorac Surg 1992;54:669–675. [DOI] [PubMed] [Google Scholar]
- 16. Knight CS, Tallaj JA, Rayburn BK, Pamboukian SV, Bourge R, Kirklin JK, et al. Bradycardia and syncope as a presentation of cardiac allograft rejection involving the conducting system. Cardiovasc Pathol 2010;19:117–120. [DOI] [PubMed] [Google Scholar]
- 17. Billingham M. A working formulation for the standardization of nomenclature in the diagnosis of heart and lung rejection: heart rejection study group. J Heart Transplant 1990;9:587–593. [PubMed] [Google Scholar]
- 18. Stewart S, Winters GL, Fishbein MC, Tazelaar HD, Kobashigawa J, Abrams J, et al. Revision of the 1990 working formulation for the standardization of nomenclature in the diagnosis of heart rejection. J Heart Lung Transplant 2005;24:1710–1720. [DOI] [PubMed] [Google Scholar]
- 19. Colvin MM, Cook JL, Chang P, Francis G, Hsu DT, Kiernan MS, et al. Antibody-mediated rejection in cardiac transplantation: emerging knowledge in diagnosis and management. Circulation 2015;131:1608–1639. [DOI] [PubMed] [Google Scholar]
- 20. Sauver J S, Buntrock J, Rademacher D. Comparison of mayo clinic coding systems. Technical Report; 2010. [Google Scholar]
- 21. Sauver JL S, Grossardt BR, Finney Rutten LJ, Roger VL, Majerus M, Jensen DW, et al. Rochester Epidemiology project data exploration portal. Prev Chronic Dis 2018;15:E42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shin H-C, Roth HR, Gao M, Lu L, Xu Z, Nogues I, et al. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging 2016;35:1285–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 1988;44:837–845. [PubMed] [Google Scholar]
- 24. Sun X, Xu W. Fast implementation of DeLong’s algorithm for comparing the areas under correlated receiver operating characteristic curves. IEEE Signal Process Lett 2014;21:1389–1393. [Google Scholar]
- 25. R Core Team R . R: A language and environment for statistical computing. R foundation for statistical computing. Vienna, Austria; 2013.
- 26. Hashim HT, Ramadhan MA, Ahmad S, Shah J, Varney J, Motawea KR, et al. The role of the electrocardiogram in the recognition of cardiac transplant rejection: a systematic review and meta-analysis. Clin Cardiol 2022;45:258–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Doering LV, Hickey K, Pickham D, Chen B, Drew BJ. Remote noninvasive allograft rejection monitoring for heart transplant recipients: study protocol for the novel evaluation with home electrocardiogram and remote transmission (NEW HEART) study. BMC Cardiovasc Disord 2012;12:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim J-YV, Lee B, Koitsopoulos P, Shannon CP, Chen V, Hollander Z, et al. Analytical validation of HEARTBiT: a blood-based Multiplex gene expression profiling assay for exclusionary diagnosis of acute cellular rejection in heart transplant patients. Clin Chem 2020;66:1063–1071. [DOI] [PubMed] [Google Scholar]
- 29. Identify Heart Transplant Injury with HeartCare. https://caredx.com/products-and-services/transplant-services/heart/heartcare/ (23 August 2022).
- 30. De Vlaminck I, Valantine HA, Snyder TM, Strehl C, Cohen G, Luikart H, et al. Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection. Sci Transl Med 2014;6:241ra77–241ra77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Agbor-Enoh S, Shah P, Tunc I, Hsu S, Russell S, Feller E, et al. Cell-free DNA to detect heart allograft acute rejection. Circulation 2021;143:1184–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim PJ, Olymbios M, Siu A, Wever Pinzon O, Adler E, Liang N, et al. A novel donor-derived cell-free DNA assay for the detection of acute rejection in heart transplantation. J Heart Lung Transplant 2022;41:919–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rinchai D, Anguiano E, Nguyen P, Chaussabel D. Finger stick blood collection for gene expression profiling and storage of tempus blood RNA tubes. F1000Res 2016;5:1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huang LH, Lin PH, Tsai KW, Wang LJ, Huang YH, Kuo HC, et al. The effects of storage temperature and duration of blood samples on DNA and RNA qualities. PLoS One 2017;12:e0184692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pérez-Carrillo L, Sánchez-Lázaro I, Triviño JC, Feijóo-Bandín S, Lago F, González-Juanatey JR, et al. Diagnostic value of serum miR-144-3p for the detection of acute cellular rejection in heart transplant patients. J Heart Lung Transplant 2022;41:137–147. [DOI] [PubMed] [Google Scholar]
- 36. Van Huyen JP D, Tible M, Gay A, Guillemain R, Aubert O, Varnous S, et al. MicroRNAs as non-invasive biomarkers of heart transplant rejection. Eur Heart J 2014;35:3194–3202. [DOI] [PubMed] [Google Scholar]
- 37. Lipkova J, Chen TY, Lu MY, Chen RJ, Shady M, Williams M, et al. Deep learning-enabled assessment of cardiac allograft rejection from endomyocardial biopsies. Nat Med 2022;28:575–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Peyster EG, Arabyarmohammadi S, Janowczyk A, Azarianpour-Esfahani S, Sekulic M, Cassol C, et al. An automated computational image analysis pipeline for histological grading of cardiac allograft rejection. Eur Heart J 2021;42:2356–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Glass C, Davis R, Xiong B, Dov D, Glass M. The use of artificial intelligence (AI) machine learning to determine myocyte damage in cardiac transplant acute cellular rejection. J Heart Lung Transplant 2020;39:S59. [Google Scholar]
- 40. Crespo-Leiro MG, Zuckermann A, Bara C, Mohacsi P, Schulz U, Boyle A, et al. Concordance among pathologists in the second cardiac allograft rejection gene expression observational study (CARGO II). Transplantation 2012;94:1172–1177. [DOI] [PubMed] [Google Scholar]
- 41. Attia ZI, Kapa S, Lopez-Jimenez F, McKie PM, Ladewig DJ, Satam G, et al. Screening for cardiac contractile dysfunction using an artificial intelligence-enabled electrocardiogram. Nat Med 2019;25:70–74. [DOI] [PubMed] [Google Scholar]
- 42. Attia ZI, Noseworthy PA, Lopez-Jimenez F, Asirvatham SJ, Deshmukh AJ, Gersh BJ, et al. An artificial intelligence-enabled ECG algorithm for the identification of patients with atrial fibrillation during sinus rhythm: a retrospective analysis of outcome prediction. Lancet 2019;394:861–867. [DOI] [PubMed] [Google Scholar]
- 43. Ko WY, Siontis KC, Attia ZI, Carter RE, Kapa S, Ommen SR, et al. Detection of hypertrophic cardiomyopathy using a convolutional neural network-enabled electrocardiogram. J Am Coll Cardiol 2020;75:722–733. [DOI] [PubMed] [Google Scholar]
- 44. Adedinsewo D, Carter RE, Attia Z, Johnson P, Kashou AH, Dugan JL, et al. Artificial intelligence-enabled ECG algorithm to identify patients with left ventricular systolic dysfunction presenting to the emergency department with dyspnea. Circ Arrhythm Electrophysiol 2020;13:e008437. [DOI] [PubMed] [Google Scholar]
- 45. Noseworthy PA, Attia ZI, Brewer LC, Hayes SN, Yao X, Kapa S, et al. Assessing and mitigating bias in medical artificial intelligence: the effects of race and ethnicity on a deep learning model for ECG analysis. Circ Arrhythm Electrophysiol 2020;13:e007988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yao X, Rushlow DR, Inselman JW, McCoy RG, Thacher TD, Behnken EM, et al. Artificial intelligence-enabled electrocardiograms for identification of patients with low ejection fraction: a pragmatic, randomized clinical trial. Nat Med 2021;27:815–819. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author. All requests for raw and analysed data and related materials, excluding programming code, will be reviewed by the Mayo Clinic legal department and Mayo Clinic Ventures to verify whether the request is subject to any intellectual property or confidentiality obligations. Requests for patient-related data not included in the paper will not be considered. Any data and materials that can be shared will be released via a Material Transfer Agreement.