Fig. 4.
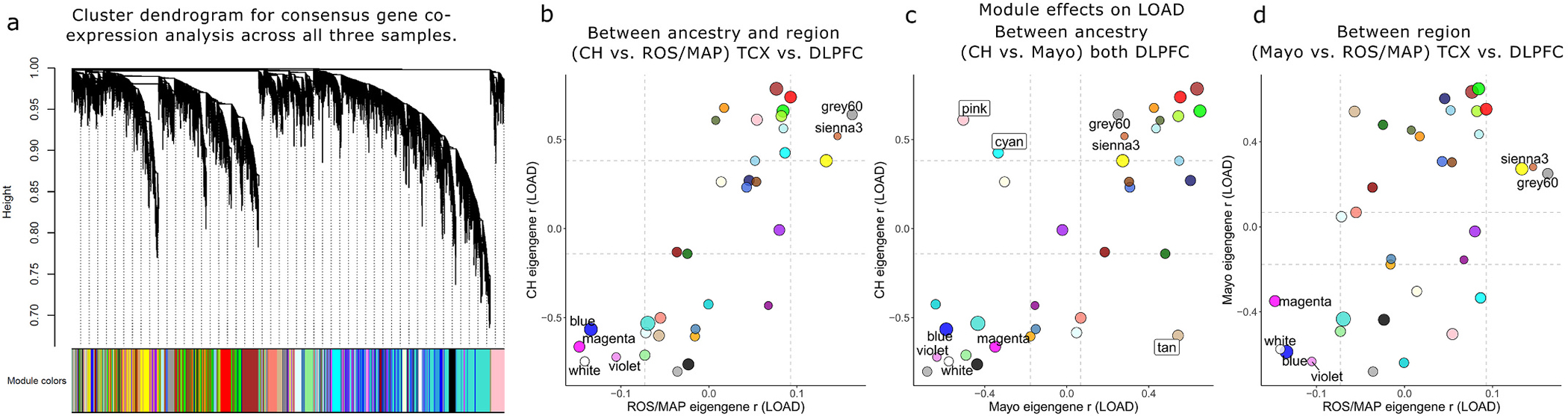
Consensus gene co-expression network module analysis with LOAD. A) Clustered dendrogram showing the common gene set hierarchical structure and consensus gene module definitions. Panels B–D show consensus module eigengene associations with LOAD (Pearson r) for all modules compared between pairwise sample combinations. Modules with concordant effects (pFDR < 0.05 with same direction of effect) in all three samples, or significantly discordant (pFDR < 0.05 in at least two samples but with opposite direction of effect), are labeled; six modules were significantly associated with LOAD in all three samples; greenyellow, brown, and royalblue modules were consistently downregulated, while the red, purple, and lightyellow modules were upregulated. In contrast, four modules showed significant but directionally discordant effects on LOAD when comparing CH to Mayo; salmon and lightcyan modules were significantly upregulated in CH but downregulated in Mayo, whereas black and cyan modules were downregulated in CH but upregulated in Mayo. Colors correspond to module definitions. CH = Caribbean-Hispanic; DLPFC = dorsolateral prefrontal cortex; ROS/MAP = Religious Orders Study and Memory and Aging Project; TCX = temporal cortex.
