Fig. 5.
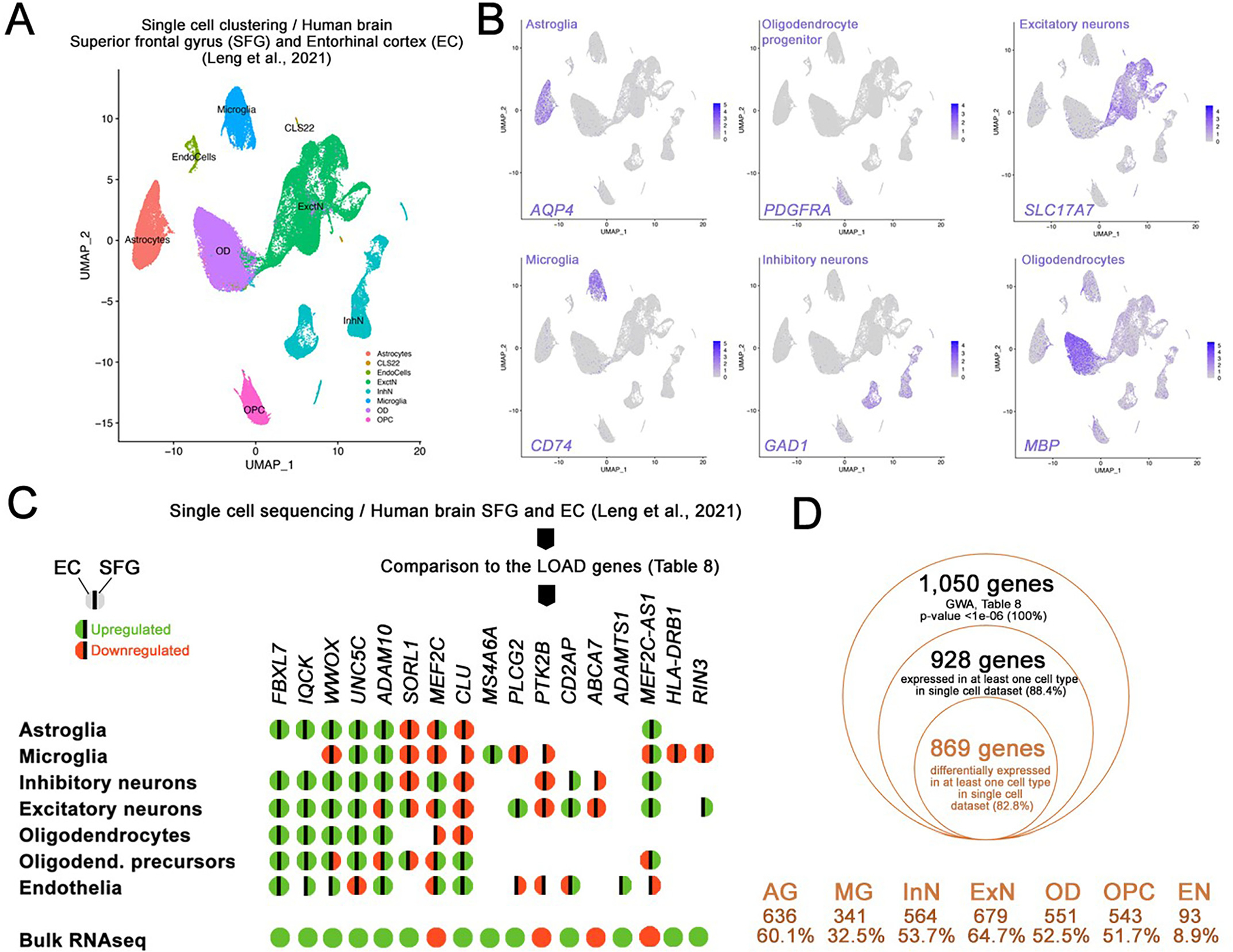
Secondary independent validation of the bulk RNA sequencing with single cell transcriptomics from human brains. (A) Clustering of single cell dataset in Leng et al., 2021 (human brains, EC and SFG, AD cases versus controls; Braak stage 6 versus 0). 7 major cell types were identified as indicated. (B) tSNE plots for major cell type markers (only one out of many were shown) AQP4 for astroglia, PDGFRA for oligodendrocyte progenitors, CD74 for microglia, GAD1 for inhibitory neurons, MBP for oligodendrocytes, SLC17A7 for excitatory neurons. (C) Genes in Table 8 were run against the single cell datasets and differentially expressed genes in various cell types in entorhinal cortex (EC) and superior frontal gyrus (SFG) of human brains with and without 17 CE genes are shown for their differential gene expression in major cell types. Green: upregulation, red: downregulation, left hemisphere: EC, right hemisphere: SFG. No color indicates no significant differential expression. (D) Comparison of GWA genes (Table 8) in single cell dataset (human EC, Braak stage 6 vs 0). 869 genes (82.6% of the initially selected GWA genes) were validated for their differential expression in human EC in at least 1 cell type. Cell types and the number/percentage of the initially selected GWA genes are shown for astroglia (AG), microglia (MG), inhibitory neurons (InN), excitatory neurons (ExN), oligodendrocytes (OD), oligodendrocyte progenitors (OPC), and endothelia (EN).
