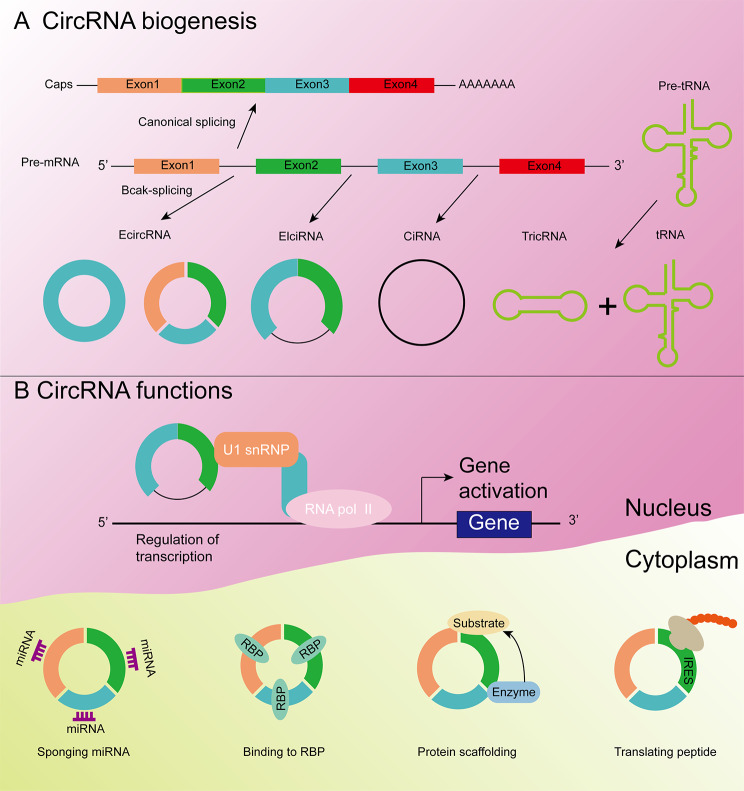
Fig. 1.
Biogenesis and function of circular RNA (circRNA)
Pre-mRNA can undergo either canonical splicing to generate a linear RNA with exon inclusion or back-splicing to produce both a circular RNA and an alternatively spliced linear RNA with exon exclusion. (A) The biogenesis of circRNAs. Biogenesis of circRNAs generates three types of circRNAs: EcircRNAs, ciRNAs, and EIciRNAs. The formation of ecircRNAs is usually by direct back splicing. Direct back splicing is a common shearing method in which precursor-mRNA is looped by linking the 5′ splice donor site downstream of the exon to the 3′ splice acceptor site upstream via covalent bonding. EIcircRNAs also use the method of direct back splicing, but in the process of back splicing, some intron sequences are not removed but remain within the circRNA. The formation of ciRNAs is thought to result from lasso formation from introns removed during pre-mRNA splicing. TricRNA is a circular RNA produced by tRNA precursor splicing. (B) The function of circRNAs. CircRNAs enhance the transcription and splicing of their parental genes by interacting with RNA pol II or U1 small nuclear ribonucleoproteins (U1 snRNPs). CircRNAs have basic functions as microRNA (miRNA) sponges, RNA-binding proteins (RBP), and templates for regulating transcription and protein translation. CircRNAs can function as microRNA sponges or decoys, protecting target mRNAs from miRNA-dependent degradation. circRNAs containing RBP binding motifs may function as sponges or decoys for these proteins and indirectly regulate their functions. circRNAs have been shown to function as protein scaffolds, facilitating the colocalization of enzymes and their substrates. CircRNA with internal ribosomal entry site elements may be translated under certain circumstances to produce unique peptides