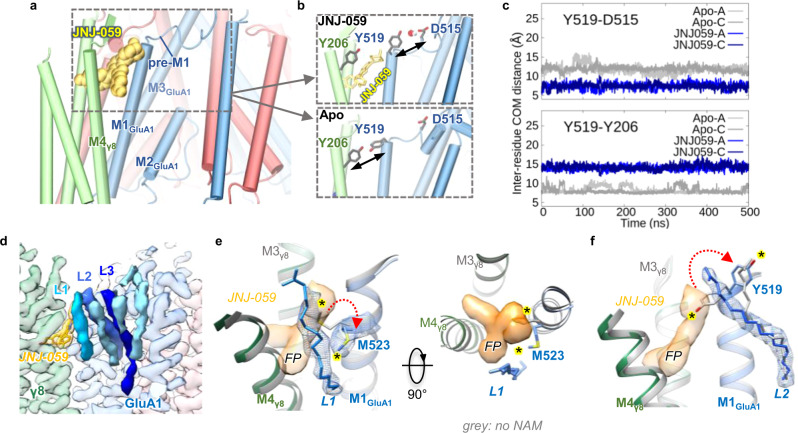
Fig. 3. JNJ-059-induced H-bond formation, and lipid interactions.
a Model of the receptor complex with JNJ-059 (yellow spheres), highlighting the proximity between the NAM pocket and the GluA1 pre-M1 helix (the figure is derived from an MD simulation). b JNJ-059 disrupts Tyr519 interaction with Tyr206 (γ8) (top panel), as compared to a simulation without the NAM (‘Apo’; bottom panel), but enables an H-bond between Tyr519 and Asp515 in the pre-M1 helix (top). c Variations in distance between centres of mass for Tyr519-Asp515 and Tyr519-Tyr206 in MD simulations of resting-state models. Distances are compared for the apo state (grey) and the JNJ059-bound structure (blue). Data are for both GluA1 subunits (chains A and C) shown in light and dark colours. d Cryo-EM map of the resting-state JNJ-059 structure. Lipid densities stacking along the GluA1 pre-M1 helix are shown in shades of blue (L1-3). e, f Overlay between the JNJ-059-resting model (coloured) with an apo resting state (PDB: 7OCD; grey). e The NAM induces an interaction between Met523 and the acyl chain of lipid L1 (side view, left; top view right), and a rearrangement of L2 via the Tyr519 side chain (f). NAM-induced side chain reorientations are highlighted with an asterisk. The flurophenyl group interacting with L1 is denoted ‘FP’. Note the dilation of the NAM pocket in the presence of JNJ-059.